6 Assignment Two
Assignment Two: The Week Three Assignment
- You are supposed to submit an R Notebook File with a file name a2_YourID.nb.html.
- Some submitted an HTML file, such as a2_YourID.html. You need to create an R Notebook. Use the template in Moodle. It creates a file with *.nb.html at the end automatically.
- Some did not run each code chunk. You should run each code or select ‘Run all’ under ‘Run’ button. If some code chunk has a problem or an error, run each code chunk or use Run all chunk above or Run all chunk below, so the result appear in your R Notebook file.
- You are supposed to write observations.
- Writing codes seem to be challenging, however, we are learning ‘data analysis’ not ‘programming’. Do not forget to write explanations of the data, questions and observations.
- Cheat Sheets, Posit Primers, and the textbook ‘R for Data Science’ are the first set of references you should look at together wih my lecture materials.
6.1 Set up
library(tidyverse)
#> ── Attaching core tidyverse packages ──── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.3 ✔ readr 2.1.4
#> ✔ forcats 1.0.0 ✔ stringr 1.5.0
#> ✔ ggplot2 3.4.4 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.3 ✔ tidyr 1.3.0
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library(gapminder)The following (df <- gapminder) is a short-hand of
df <- gapminder
df
(df <- gapminder)
#> # A tibble: 1,704 × 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> 4 Afghanistan Asia 1967 34.0 11537966 836.
#> 5 Afghanistan Asia 1972 36.1 13079460 740.
#> 6 Afghanistan Asia 1977 38.4 14880372 786.
#> 7 Afghanistan Asia 1982 39.9 12881816 978.
#> 8 Afghanistan Asia 1987 40.8 13867957 852.
#> 9 Afghanistan Asia 1992 41.7 16317921 649.
#> 10 Afghanistan Asia 1997 41.8 22227415 635.
#> # ℹ 1,694 more rows6.2 General Comments
6.2.1 Varibles
We should know first about the variables. At least you must know if each variable is a categorical or a numerical variable.
For example, in the gapminder data, country, continent are categorical variables, and year, lifeExp, pop, gdpPercap are numerical variables. It is possible to treat year as a categorical variable.
6.2.2 Example: datasets::CO2
6.2.2.1 The first step
You can obtain basic information about the data by the following or by typing CO2 in the search box under the Help tab. You can see the same at: https://stat.ethz.ch/R-manual/R-devel/library/datasets/html/00Index.html
help(CO2) # or ? CO2- Description: The CO2 data frame has 84 rows and 5 columns of data from an experiment on the cold tolerance of the grass species Echinochloa crus-galli.
- Usage: CO2
-
Format
An object of class c(“nfnGroupedData”, “nfGroupedData”, “groupedData”, “data.frame”) containing the following columns:
Plant: an ordered factor with levels Qn1 < Qn2 < Qn3 < … < Mc1 giving a unique identifier for each plant.
Type: a factor with levels Quebec Mississippi giving the origin of the plant
Treatment: a factor with levels nonchilled chilled
conc: a numeric vector of ambient carbon dioxide concentrations (mL/L).
uptake: a numeric vector of carbon dioxide uptake rates (/m^2μmol/m 2 sec).
-
Details: The CO_2 uptake of six plants from Quebec and six plants from Mississippi was measured at several levels of ambient CO_2 concentration. Half the plants of each type were chilled overnight before the experiment was conducted.
- This dataset was originally part of package nlme, and that has methods (including for [, as.data.frame, plot and print) for its grouped-data classes.
-
Source: Potvin, C., Lechowicz, M. J. and Tardif, S. (1990) “The statistical analysis of ecophysiological response curves obtained from experiments involving repeated measures”, Ecology, 71, 1389–1400.
- Pinheiro, J. C. and Bates, D. M. (2000) Mixed-effects Models in S and S-PLUS, Springer.
df_co2 <- as_tibble(datasets::CO2) # what happens if simply `df_co2 <- datasets::CO2`
df_co2
#> # A tibble: 84 × 5
#> Plant Type Treatment conc uptake
#> <ord> <fct> <fct> <dbl> <dbl>
#> 1 Qn1 Quebec nonchilled 95 16
#> 2 Qn1 Quebec nonchilled 175 30.4
#> 3 Qn1 Quebec nonchilled 250 34.8
#> 4 Qn1 Quebec nonchilled 350 37.2
#> 5 Qn1 Quebec nonchilled 500 35.3
#> 6 Qn1 Quebec nonchilled 675 39.2
#> 7 Qn1 Quebec nonchilled 1000 39.7
#> 8 Qn2 Quebec nonchilled 95 13.6
#> 9 Qn2 Quebec nonchilled 175 27.3
#> 10 Qn2 Quebec nonchilled 250 37.1
#> # ℹ 74 more rowsYou can use head(CO2) if you set df_co2 <-CO2 or df_co2 <- datasets::CO2.
glimpse(df_co2)
#> Rows: 84
#> Columns: 5
#> $ Plant <ord> Qn1, Qn1, Qn1, Qn1, Qn1, Qn1, Qn1, Qn2, …
#> $ Type <fct> Quebec, Quebec, Quebec, Quebec, Quebec, …
#> $ Treatment <fct> nonchilled, nonchilled, nonchilled, nonc…
#> $ conc <dbl> 95, 175, 250, 350, 500, 675, 1000, 95, 1…
#> $ uptake <dbl> 16.0, 30.4, 34.8, 37.2, 35.3, 39.2, 39.7…“factor” is a categorical data, and “double” is a numerical data.
class(df_co2$Plant)
#> [1] "ordered" "factor"
class(df_co2$Type)
#> [1] "factor"
class(df_co2$Treatment)
#> [1] "factor"
class(df_co2$conc)
#> [1] "numeric"
class(df_co2$uptake)
#> [1] "numeric"
summary(df_co2)
#> Plant Type Treatment
#> Qn1 : 7 Quebec :42 nonchilled:42
#> Qn2 : 7 Mississippi:42 chilled :42
#> Qn3 : 7
#> Qc1 : 7
#> Qc3 : 7
#> Qc2 : 7
#> (Other):42
#> conc uptake
#> Min. : 95 Min. : 7.70
#> 1st Qu.: 175 1st Qu.:17.90
#> Median : 350 Median :28.30
#> Mean : 435 Mean :27.21
#> 3rd Qu.: 675 3rd Qu.:37.12
#> Max. :1000 Max. :45.50
#> 6.2.2.2 Try as many visualizations as possible
Then you can choose appropriate ones later in your research.
6.2.2.2.1 Histogram
Can you tell why you get the chart below?
df_co2 %>% ggplot(aes(x = conc)) + geom_histogram()
#> `stat_bin()` using `bins = 30`. Pick better value with
#> `binwidth`.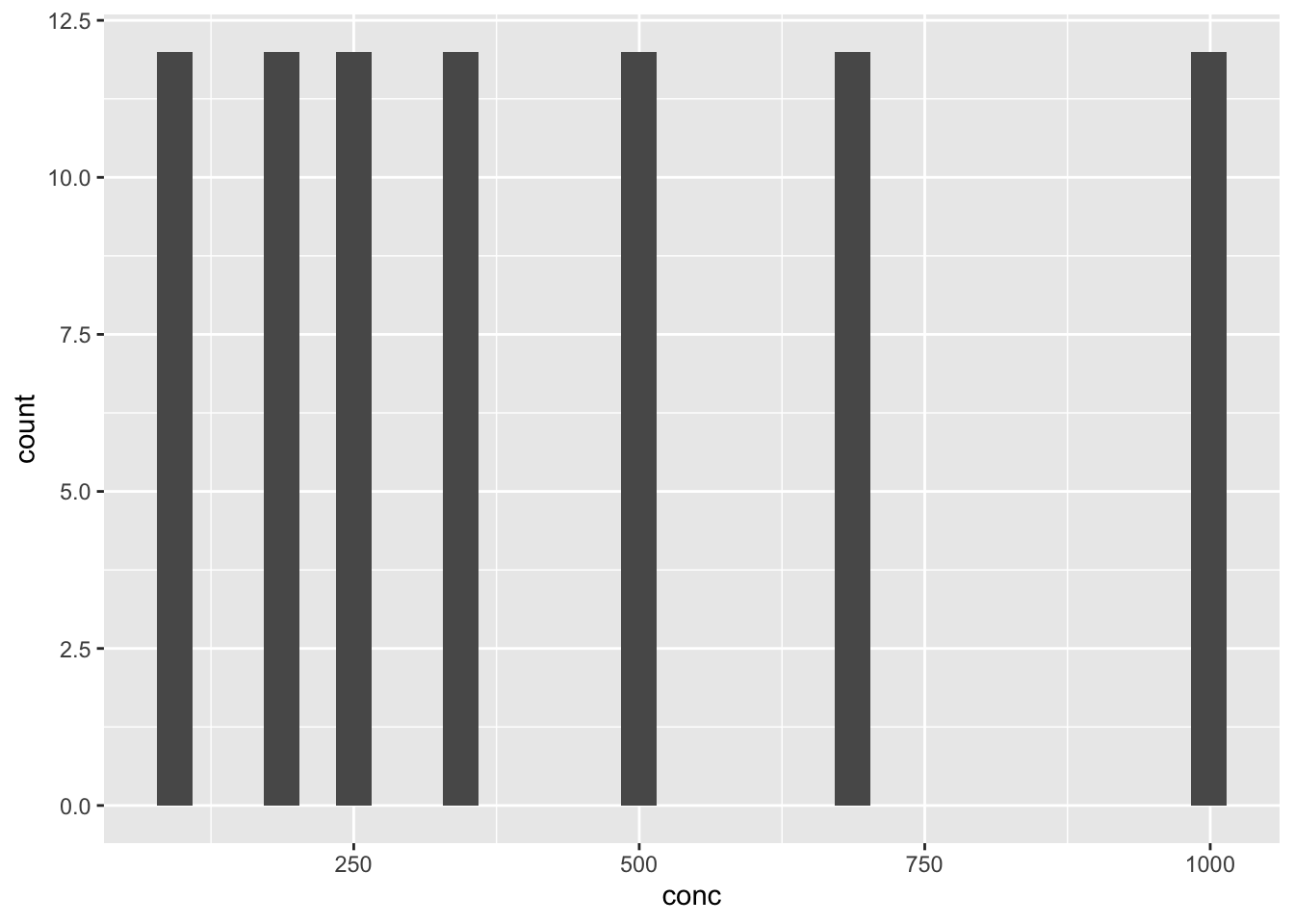
df_co2 %>% ggplot(aes(x = uptake)) + geom_histogram()
#> `stat_bin()` using `bins = 30`. Pick better value with
#> `binwidth`.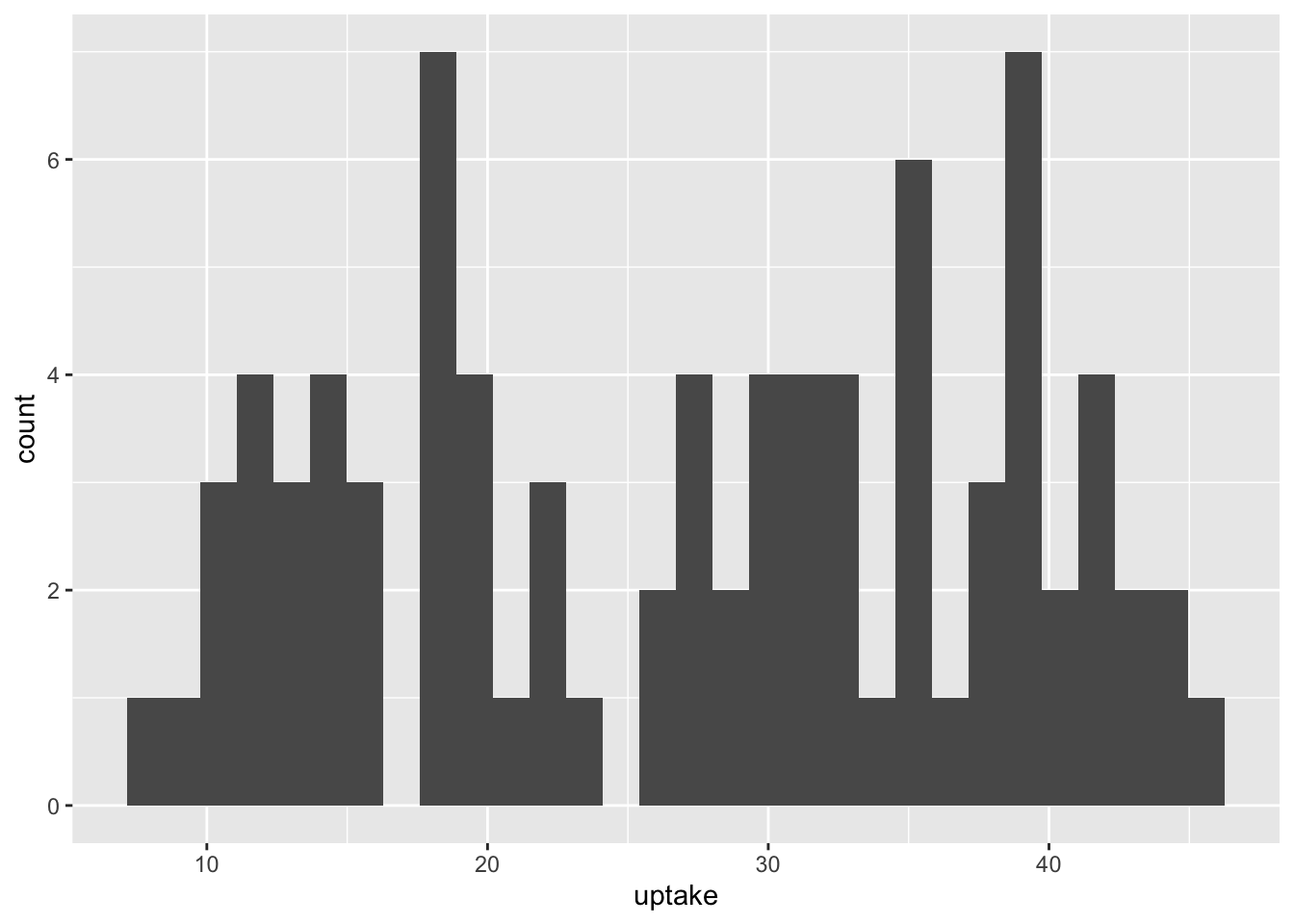
6.2.2.2.2 Box Plots
df_co2 %>% ggplot(aes(x = factor(conc), y = uptake)) + geom_boxplot()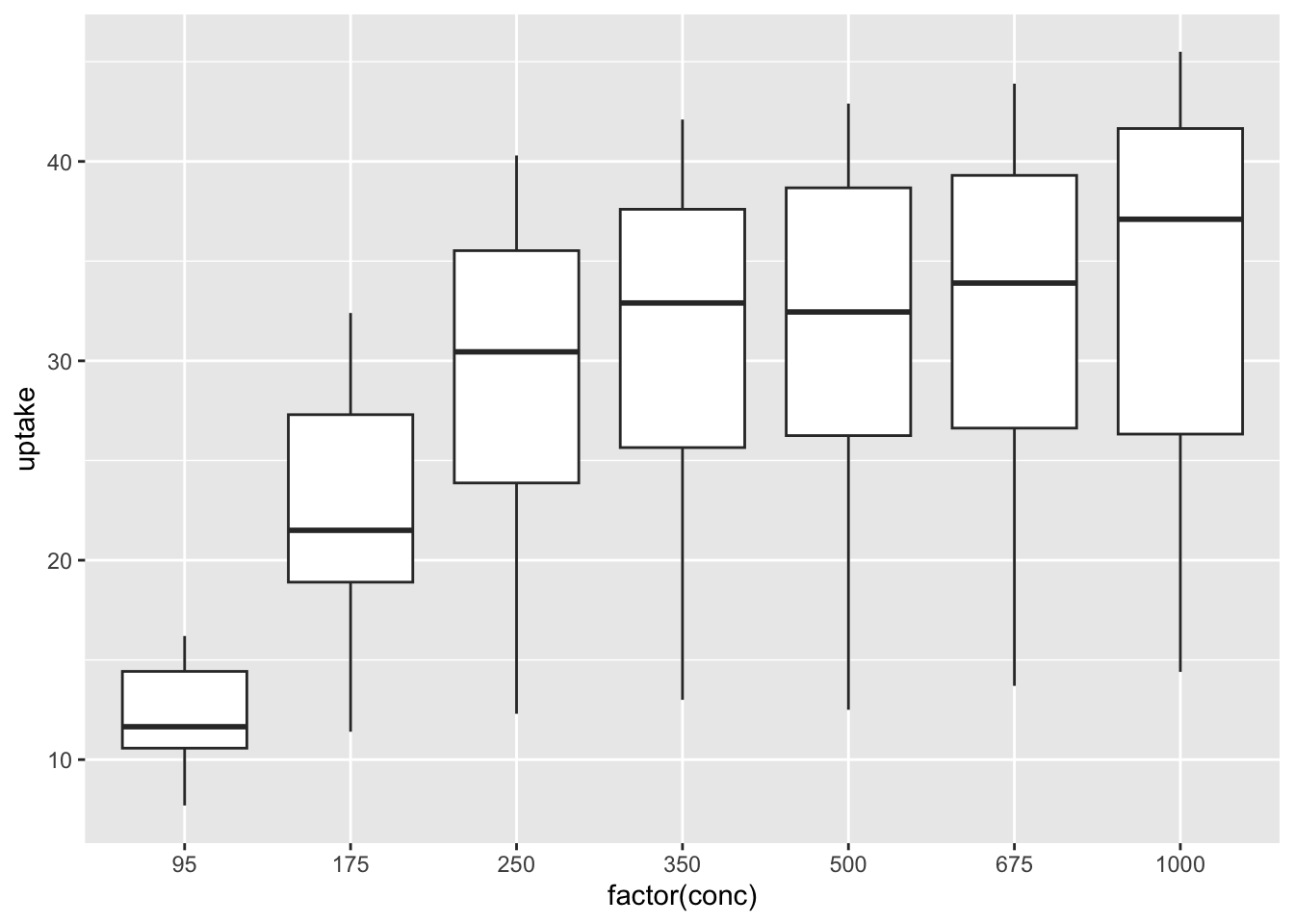
df_co2 %>% ggplot(aes(x = factor(conc), y = uptake, fill = Type)) + geom_boxplot()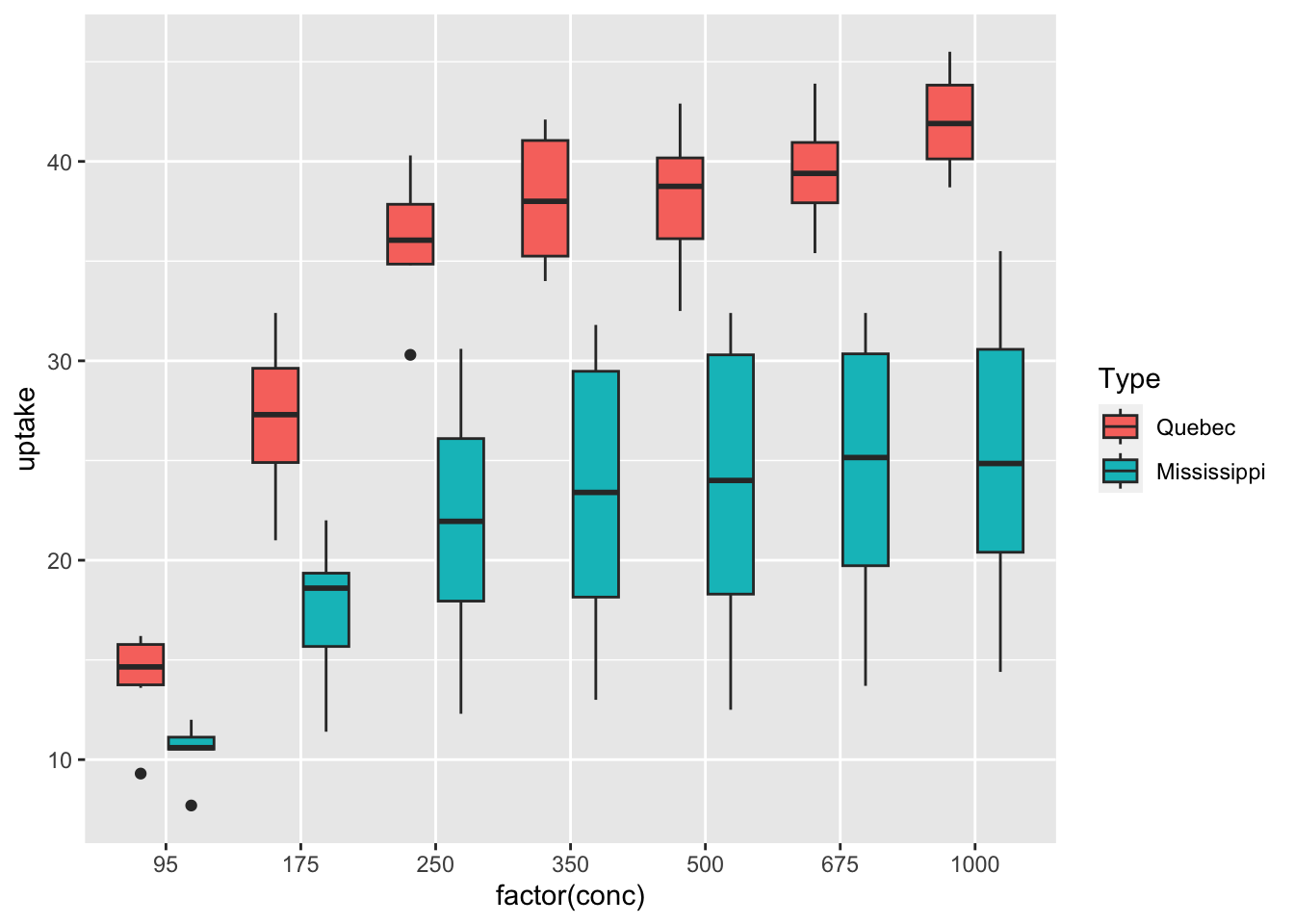
df_co2 %>% ggplot(aes(x = factor(conc), y = uptake, fill = Treatment)) + geom_boxplot()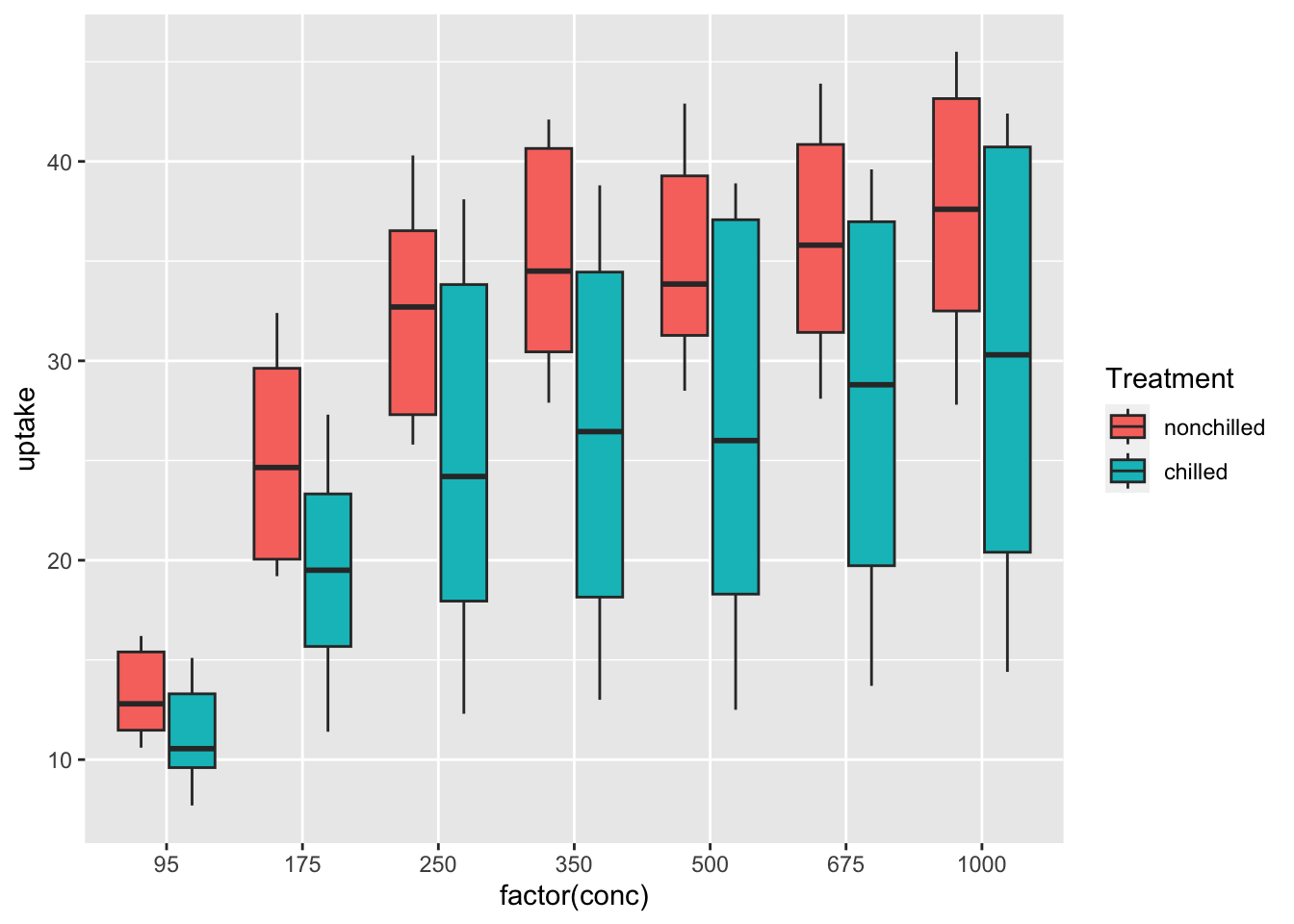
df_co2 %>% ggplot(aes(x = Plant, y = uptake, fill = Treatment)) + geom_boxplot()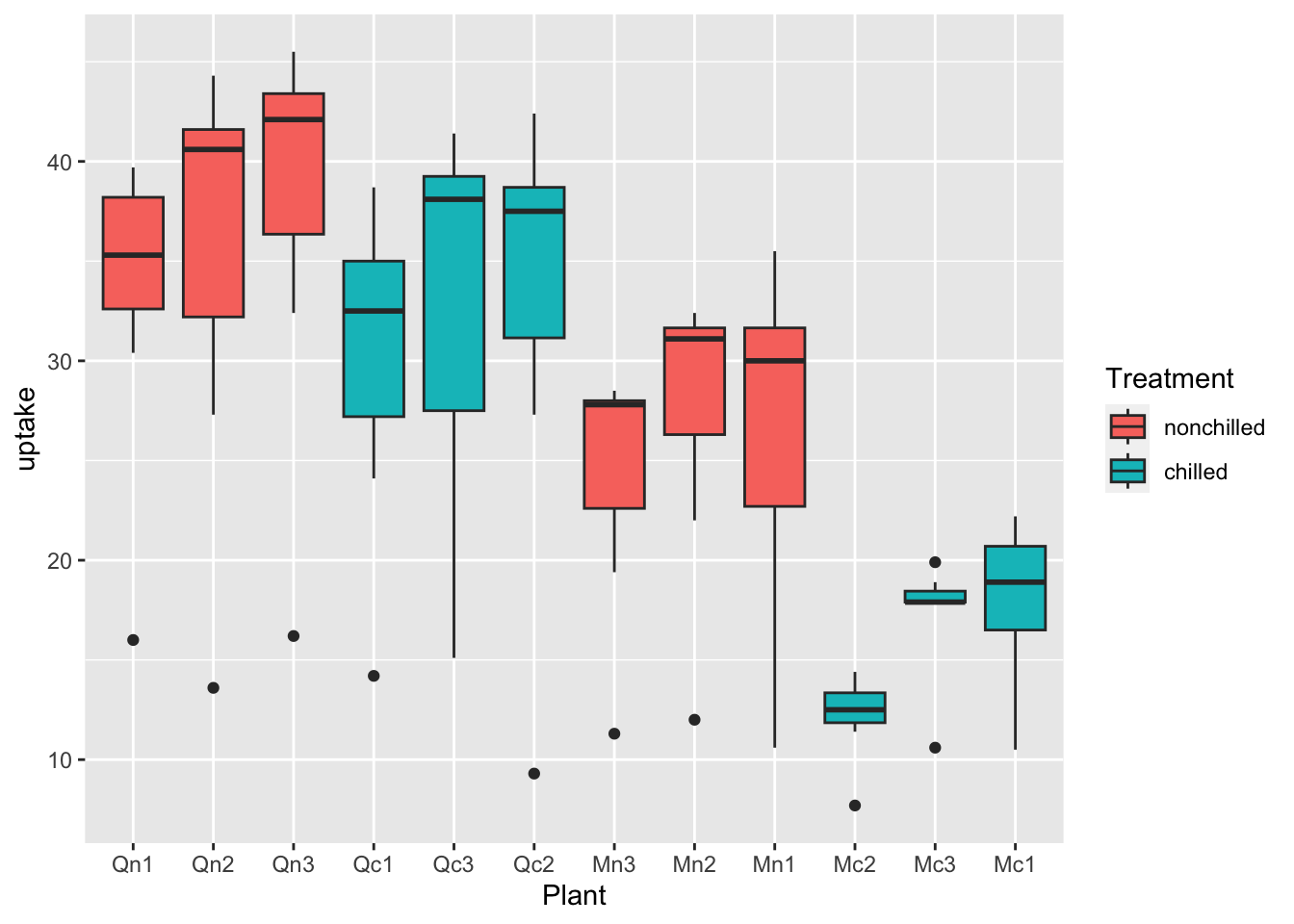
df_co2 %>% ggplot(aes(x = Plant, y = uptake, fill = Type)) + geom_boxplot()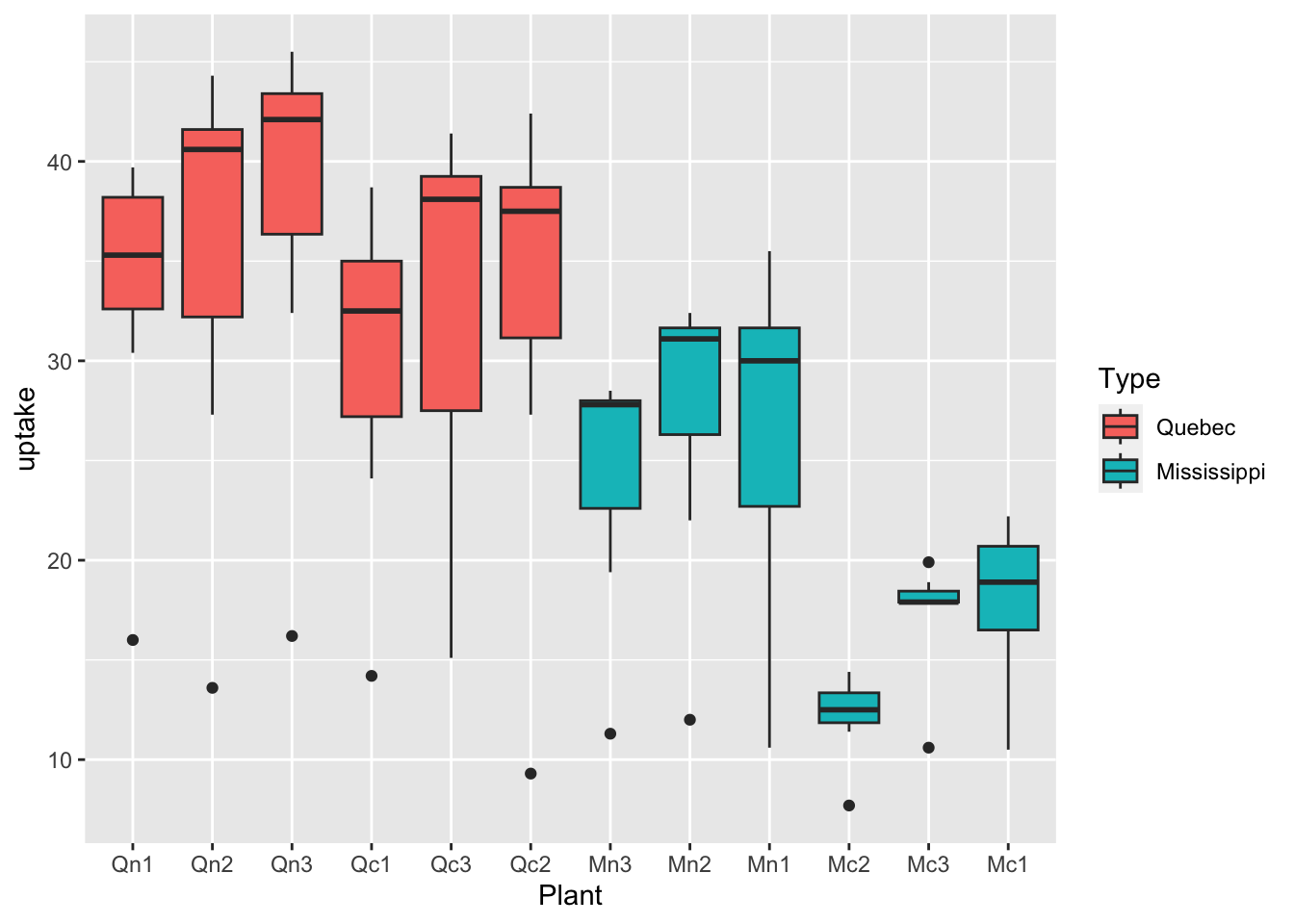
df_co2 %>% ggplot(aes(x = Plant, y = uptake, fill = Type, color = Treatment)) + geom_boxplot()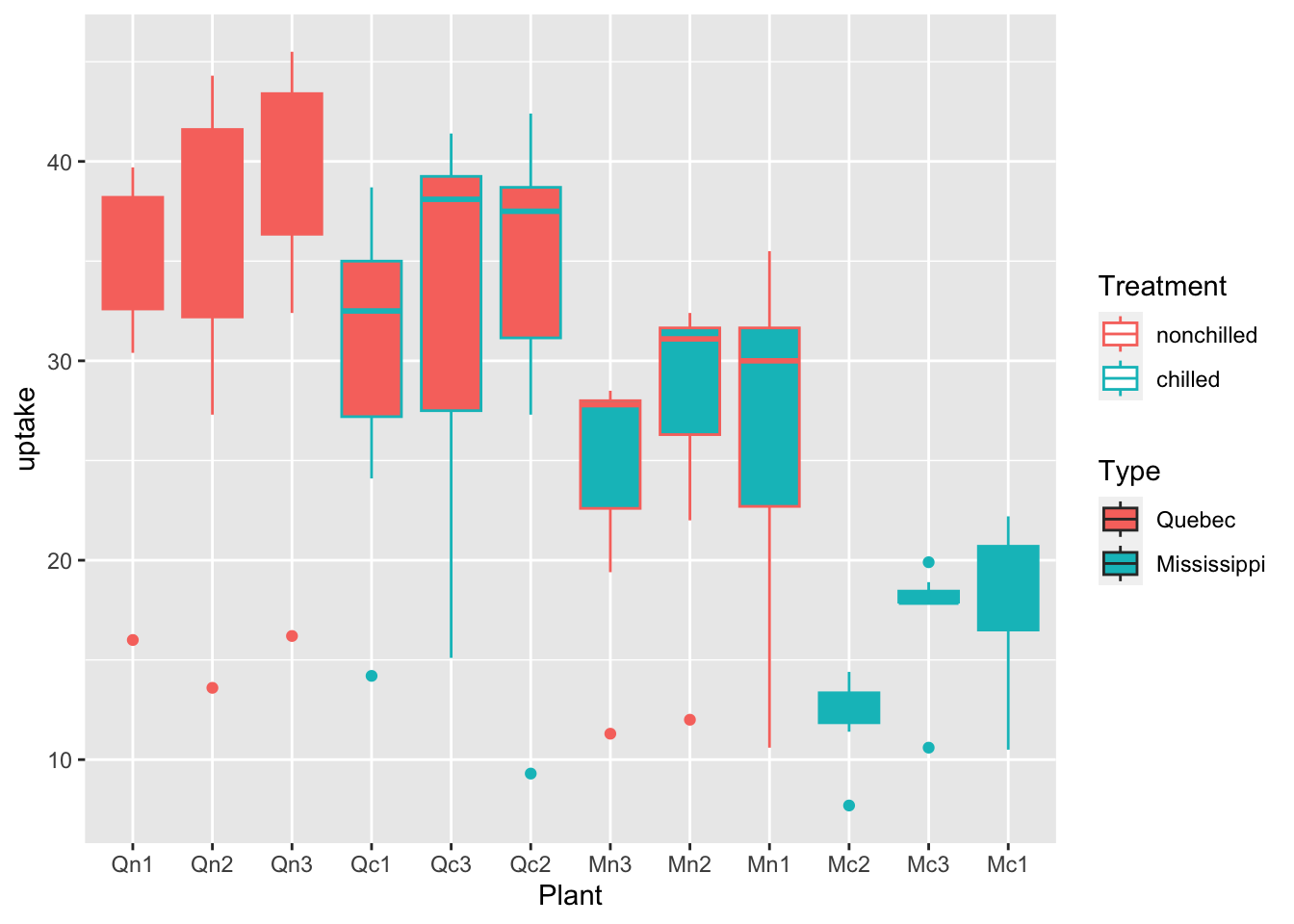
What can you see? Write your observations.
6.2.2.2.3 Scatter Plots
df_co2 %>% ggplot(aes(x = conc, y = uptake, color = Treatment)) + geom_point()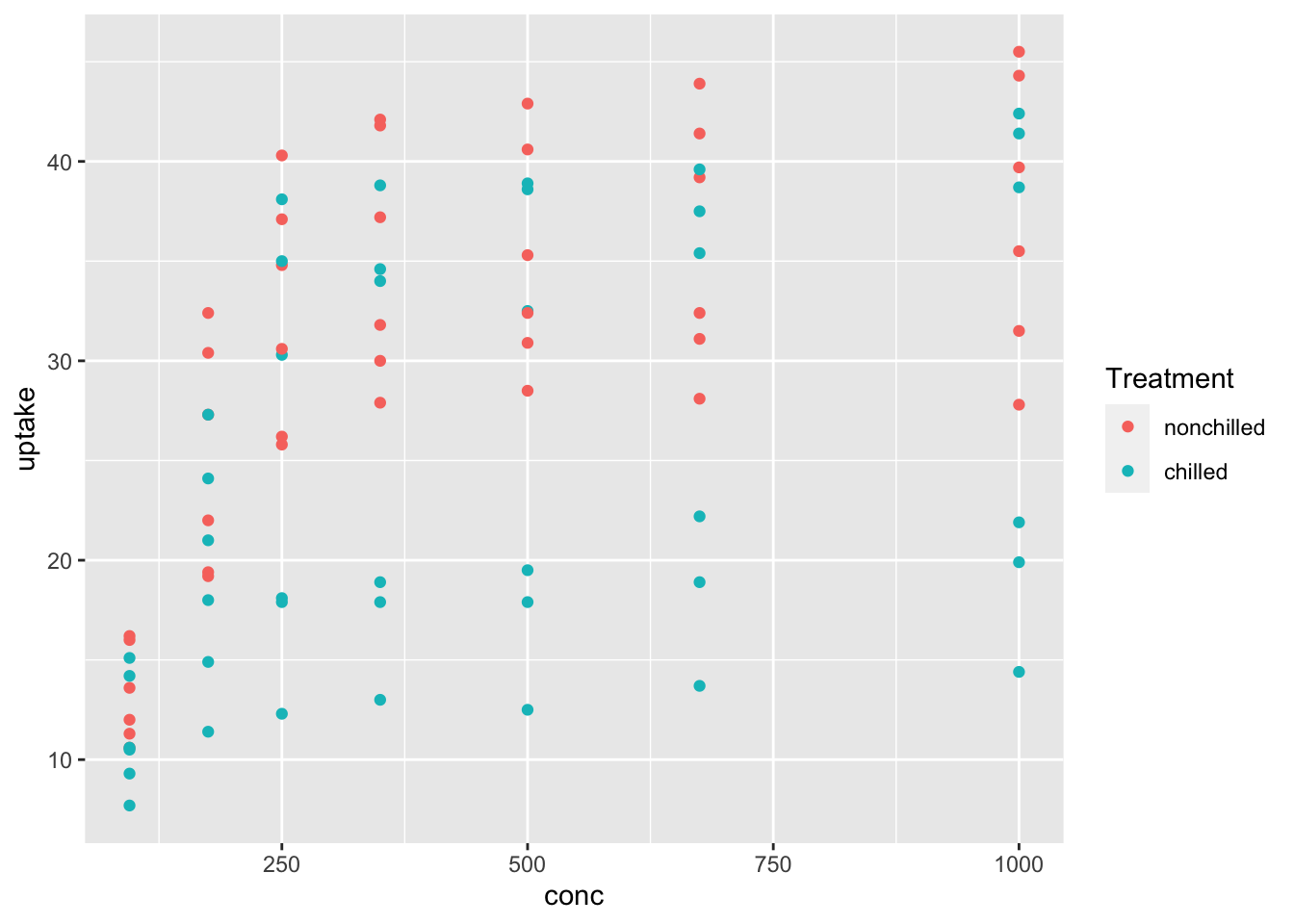
df_co2 %>% ggplot(aes(x = Plant, y = Type, color = Treatment, size = conc)) + geom_point()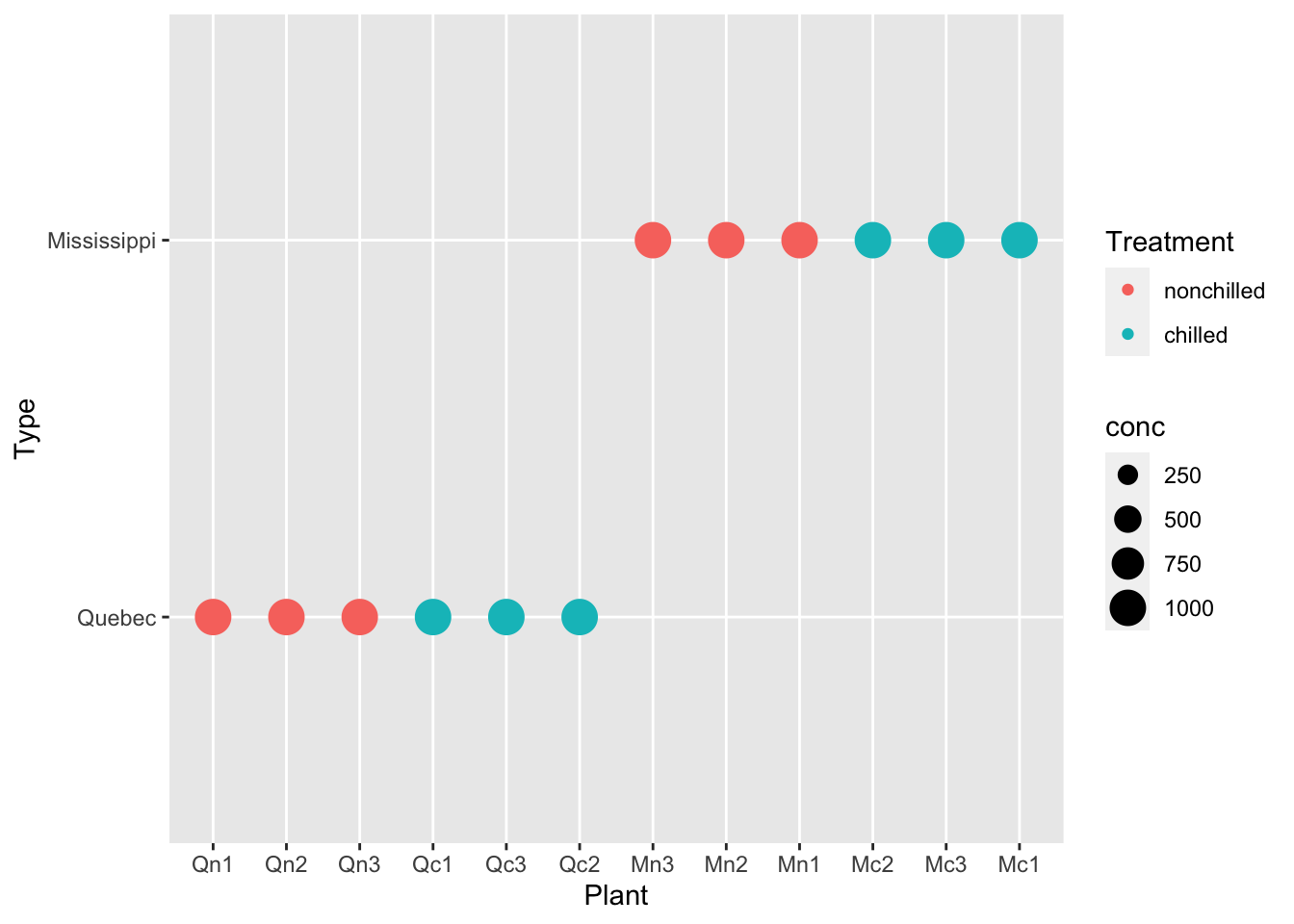
df_co2 %>% ggplot(aes(x = Plant, y = Type, size = conc, shape = Treatment)) + geom_point() + facet_wrap(vars(Treatment))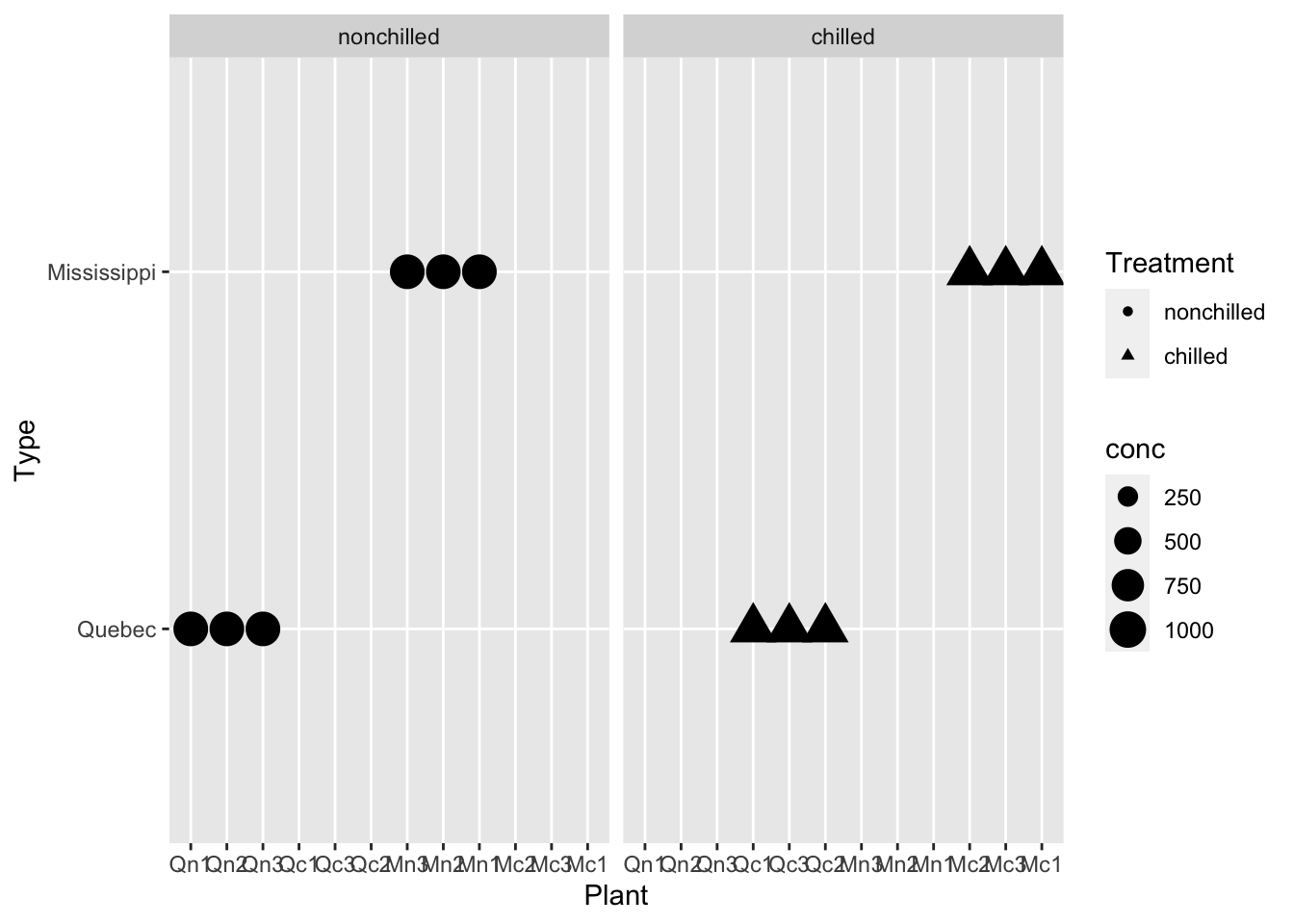
ggplot(data = df_co2) +
geom_point(aes(x = conc, y = uptake))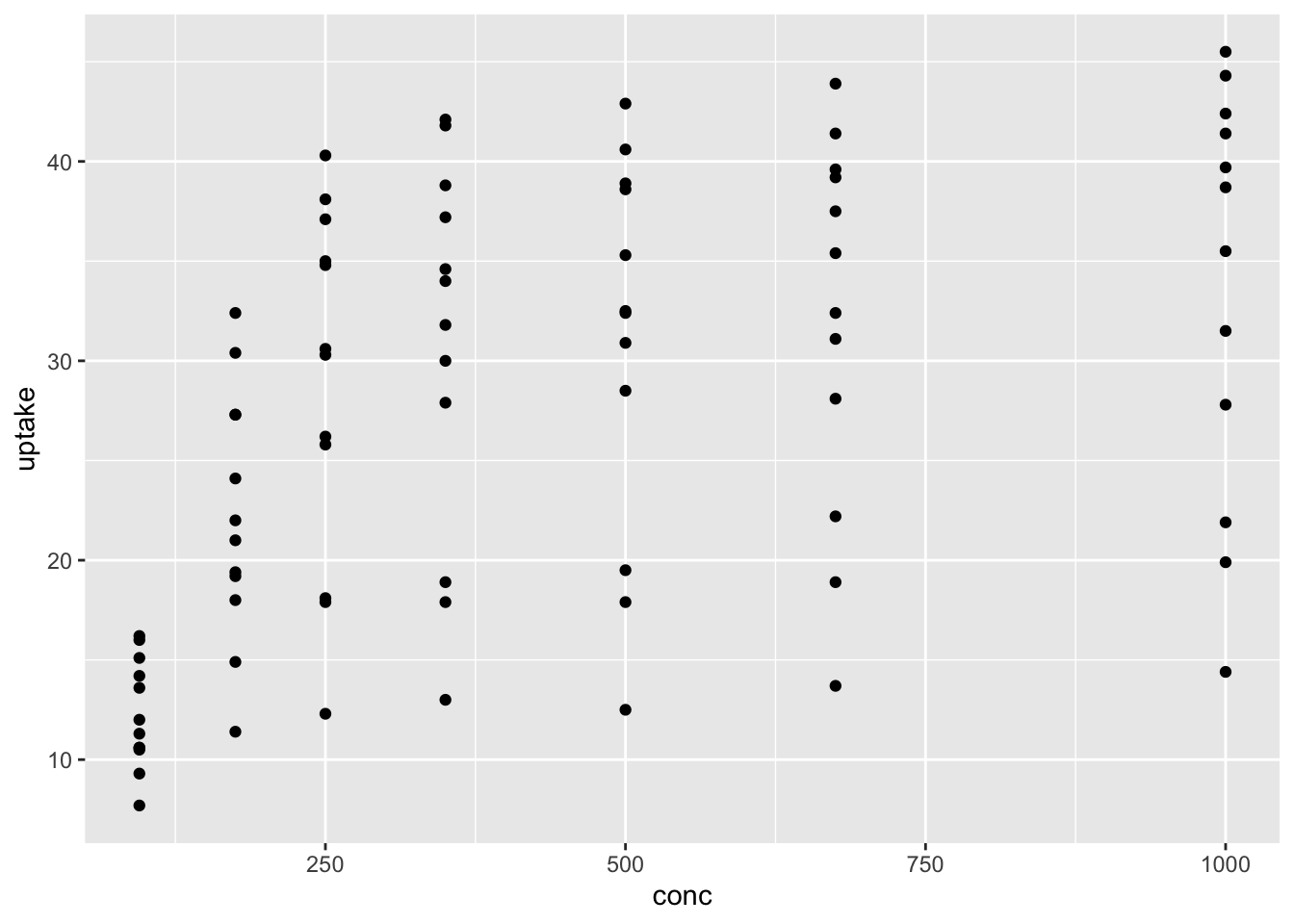
The following prints a vector.
The following code generates a data frame.
df_co2 %>% distinct(conc)
#> # A tibble: 7 × 1
#> conc
#> <dbl>
#> 1 95
#> 2 175
#> 3 250
#> 4 350
#> 5 500
#> 6 675
#> 7 1000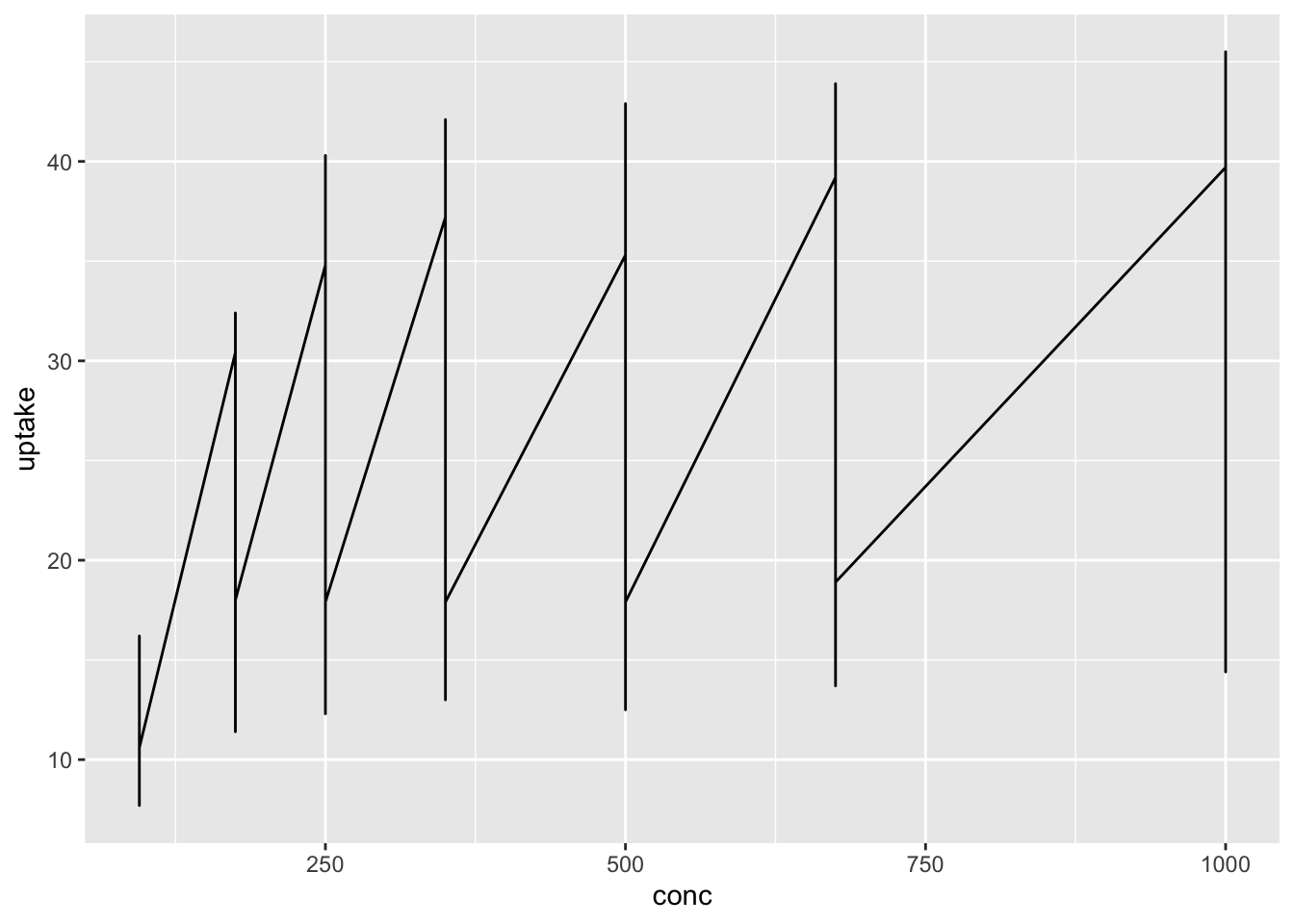
The code above did not work, and the line graph is not appropriate in this case. There are so many update values at the same conc.
6.2.2.3 Example. datasets::Seatbelts
Search the data information.
Road Casualties in Great Britain 1969–84
- Seatbelts is a multiple time series, with columns
- DriversKilled: car drivers killed.
- drivers: same as UKDriverDeaths.
- front: front-seat passengers killed or seriously injured.
- rear: rear-seat passengers killed or seriously injured.
- kms: distance driven.
- PetrolPrice: petrol price.
- VanKilled: number of van (‘light goods vehicle’) drivers.
- law: 0/1: was the law in effect that month?
References Harvey, A. C. and Durbin, J. (1986). The effects of seat belt legislation on British road casualties: A case study in structural time series modelling. Journal of the Royal Statistical Society series A, 149, 187–227. doi:10.2307/2981553.
The paper is available as you log-in to ICU Library > E-Databases > JSTOR
Can you see the difference of the following two codes?
head(Seatbelts)
#> DriversKilled drivers front rear kms PetrolPrice
#> [1,] 107 1687 867 269 9059 0.1029718
#> [2,] 97 1508 825 265 7685 0.1023630
#> [3,] 102 1507 806 319 9963 0.1020625
#> [4,] 87 1385 814 407 10955 0.1008733
#> [5,] 119 1632 991 454 11823 0.1010197
#> [6,] 106 1511 945 427 12391 0.1005812
#> VanKilled law
#> [1,] 12 0
#> [2,] 6 0
#> [3,] 12 0
#> [4,] 8 0
#> [5,] 10 0
#> [6,] 13 0
df_sb <- as_tibble(datasets::Seatbelts)
df_sb
#> # A tibble: 192 × 8
#> DriversKilled drivers front rear kms PetrolPrice
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 107 1687 867 269 9059 0.103
#> 2 97 1508 825 265 7685 0.102
#> 3 102 1507 806 319 9963 0.102
#> 4 87 1385 814 407 10955 0.101
#> 5 119 1632 991 454 11823 0.101
#> 6 106 1511 945 427 12391 0.101
#> 7 110 1559 1004 522 13460 0.104
#> 8 106 1630 1091 536 14055 0.104
#> 9 107 1579 958 405 12106 0.104
#> 10 134 1653 850 437 11372 0.103
#> # ℹ 182 more rows
#> # ℹ 2 more variables: VanKilled <dbl>, law <dbl>
summary(df_sb)
#> DriversKilled drivers front
#> Min. : 60.0 Min. :1057 Min. : 426.0
#> 1st Qu.:104.8 1st Qu.:1462 1st Qu.: 715.5
#> Median :118.5 Median :1631 Median : 828.5
#> Mean :122.8 Mean :1670 Mean : 837.2
#> 3rd Qu.:138.0 3rd Qu.:1851 3rd Qu.: 950.8
#> Max. :198.0 Max. :2654 Max. :1299.0
#> rear kms PetrolPrice
#> Min. :224.0 Min. : 7685 Min. :0.08118
#> 1st Qu.:344.8 1st Qu.:12685 1st Qu.:0.09258
#> Median :401.5 Median :14987 Median :0.10448
#> Mean :401.2 Mean :14994 Mean :0.10362
#> 3rd Qu.:456.2 3rd Qu.:17202 3rd Qu.:0.11406
#> Max. :646.0 Max. :21626 Max. :0.13303
#> VanKilled law
#> Min. : 2.000 Min. :0.0000
#> 1st Qu.: 6.000 1st Qu.:0.0000
#> Median : 8.000 Median :0.0000
#> Mean : 9.057 Mean :0.1198
#> 3rd Qu.:12.000 3rd Qu.:0.0000
#> Max. :17.000 Max. :1.0000Which visualization do you apply?
df_sb %>% ggplot(aes(x = factor(law), y = DriversKilled)) + geom_boxplot()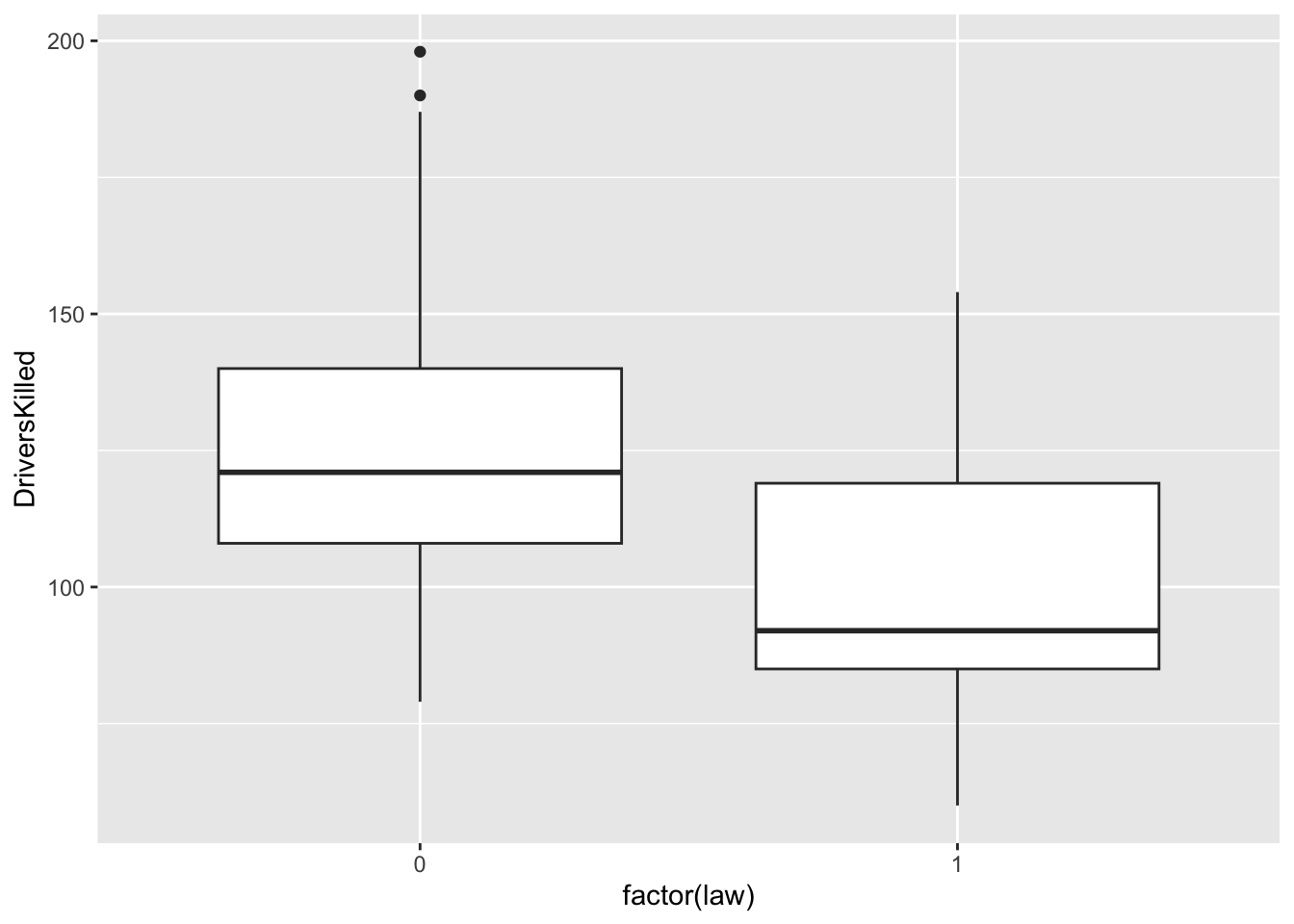
What do you observe above?
df_sb %>% ggplot(aes(x = PetrolPrice, y = DriversKilled)) + geom_point() +
geom_smooth(formula = y~x, method = "lm", se = FALSE)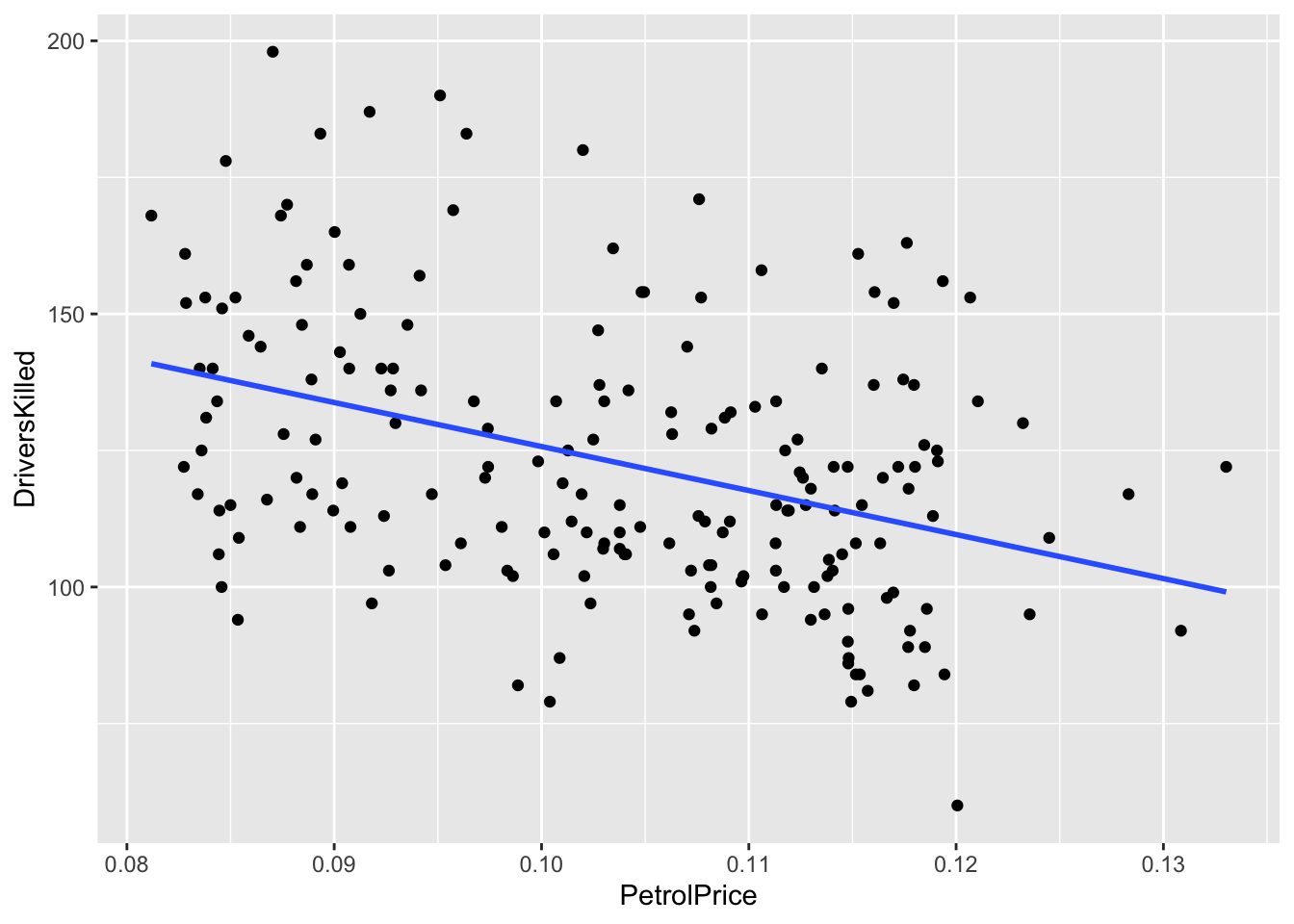
What can you see above?
df_sb %>% ggplot(aes(x = kms, y = DriversKilled)) + geom_point() +
geom_smooth(formula = y~x, method = "lm", se = FALSE)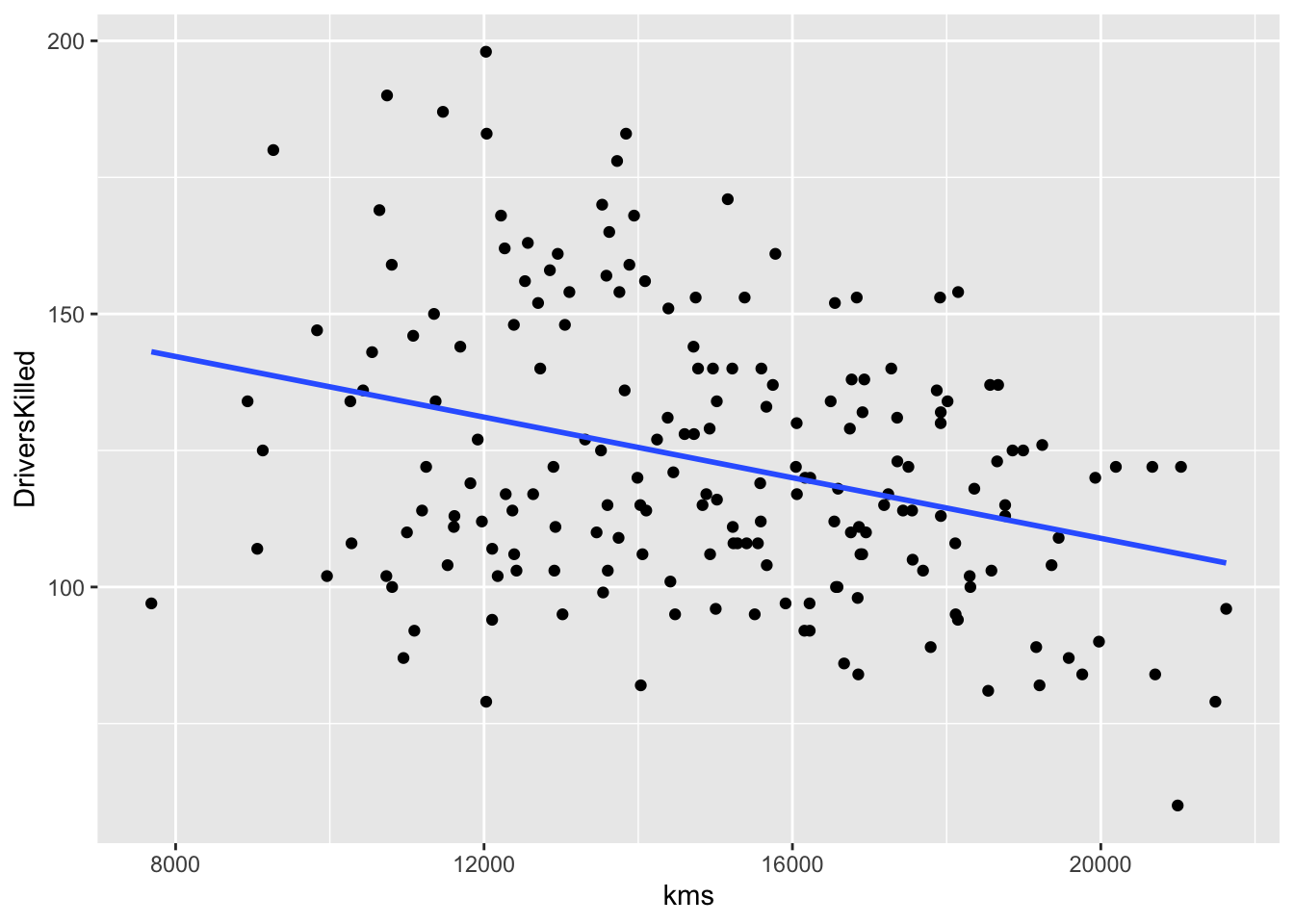
What can you see above?
We will learn how to use pivot_longer and pivot_wider in EDA4.
df_sb %>%
pivot_longer(cols = 2:4, names_to = "seat", values_to = "value")
#> # A tibble: 576 × 7
#> DriversKilled kms PetrolPrice VanKilled law seat
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 107 9059 0.103 12 0 drivers
#> 2 107 9059 0.103 12 0 front
#> 3 107 9059 0.103 12 0 rear
#> 4 97 7685 0.102 6 0 drivers
#> 5 97 7685 0.102 6 0 front
#> 6 97 7685 0.102 6 0 rear
#> 7 102 9963 0.102 12 0 drivers
#> 8 102 9963 0.102 12 0 front
#> 9 102 9963 0.102 12 0 rear
#> 10 87 10955 0.101 8 0 drivers
#> # ℹ 566 more rows
#> # ℹ 1 more variable: value <dbl>
df_sb %>%
pivot_longer(cols = 2:4, names_to = "seat", values_to = "value") %>%
ggplot() + geom_boxplot(aes(x = seat, y = value, fill = seat))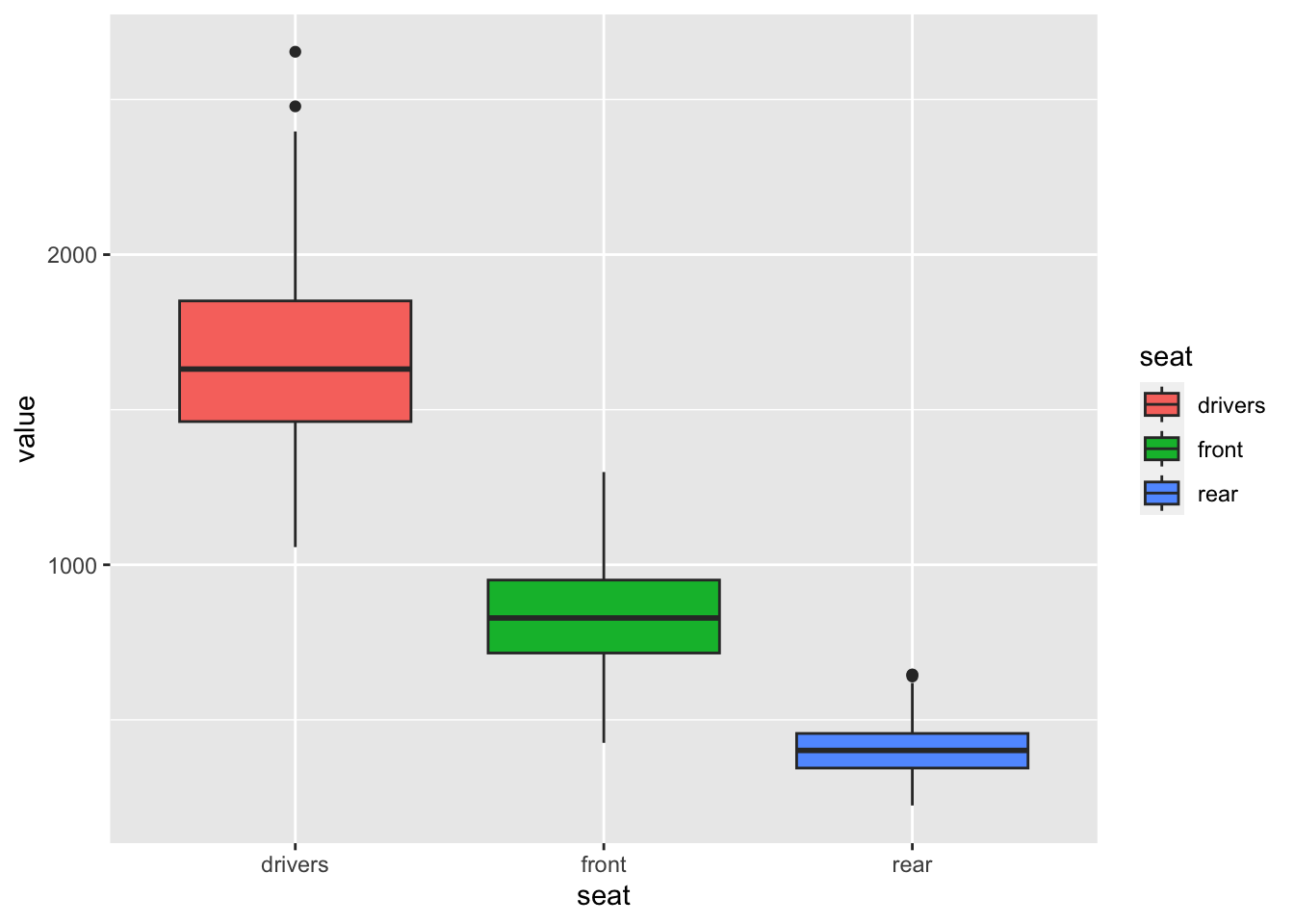
What can you observe?
df %>% filter(continent == "Asia") %>%
ggplot(aes(x = year, y = gdpPercap, color = country)) + geom_line()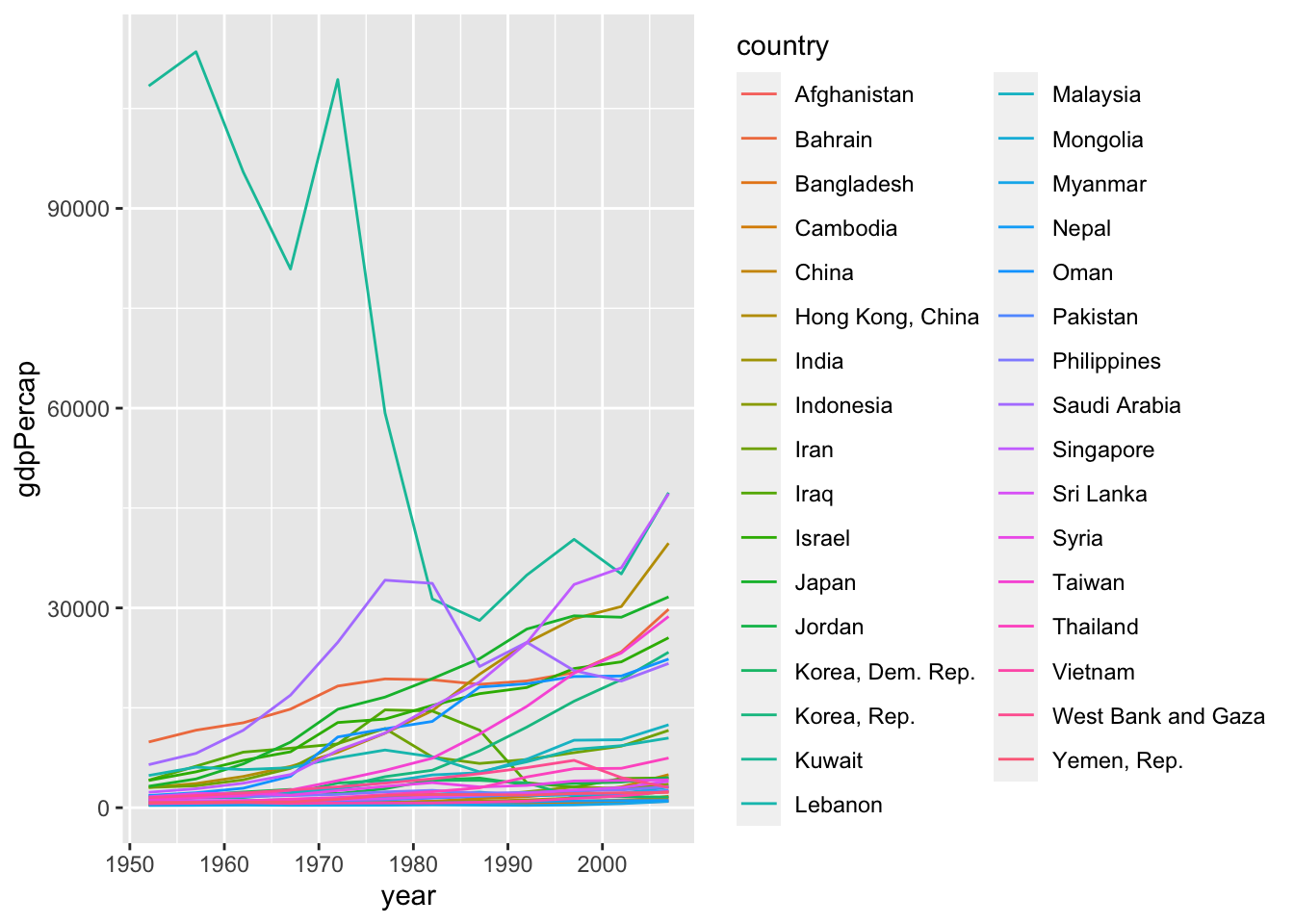
Appropriate graph?
6.2.3 Gapminder
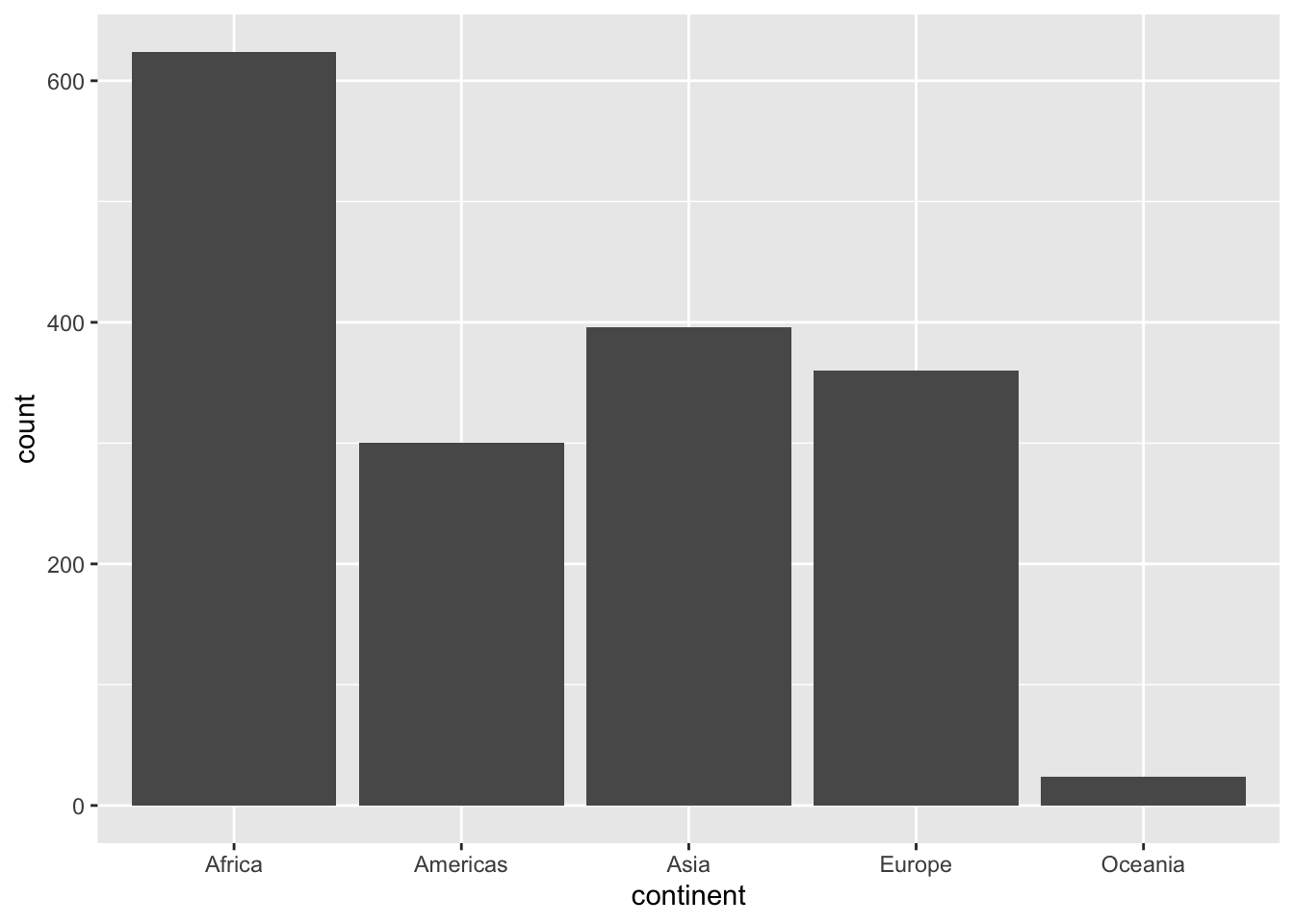
ggplot(df, aes(x = continent, y = pop)) + geom_boxplot()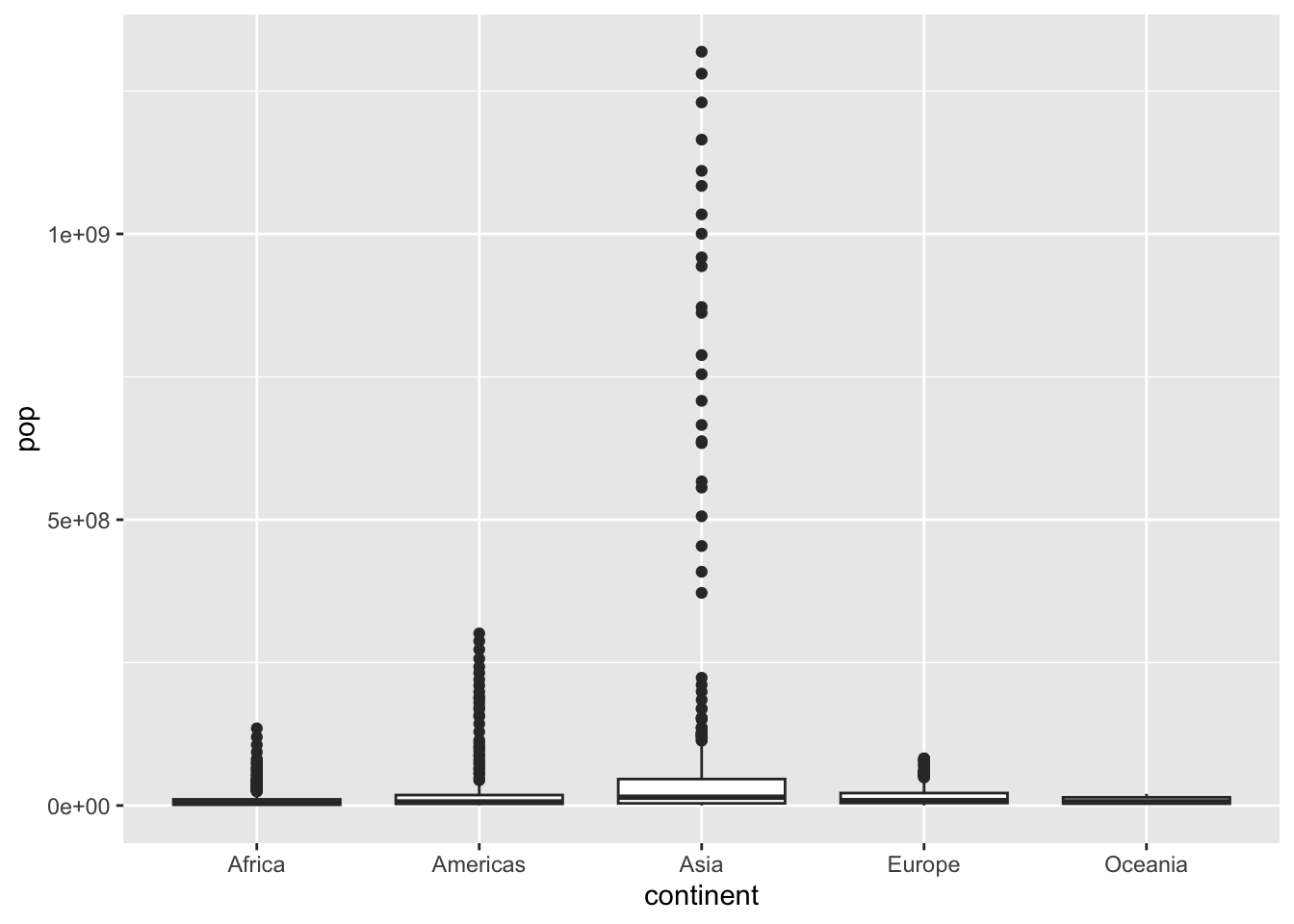
ggplot(df, aes(x = continent, y = log10(pop))) + geom_boxplot()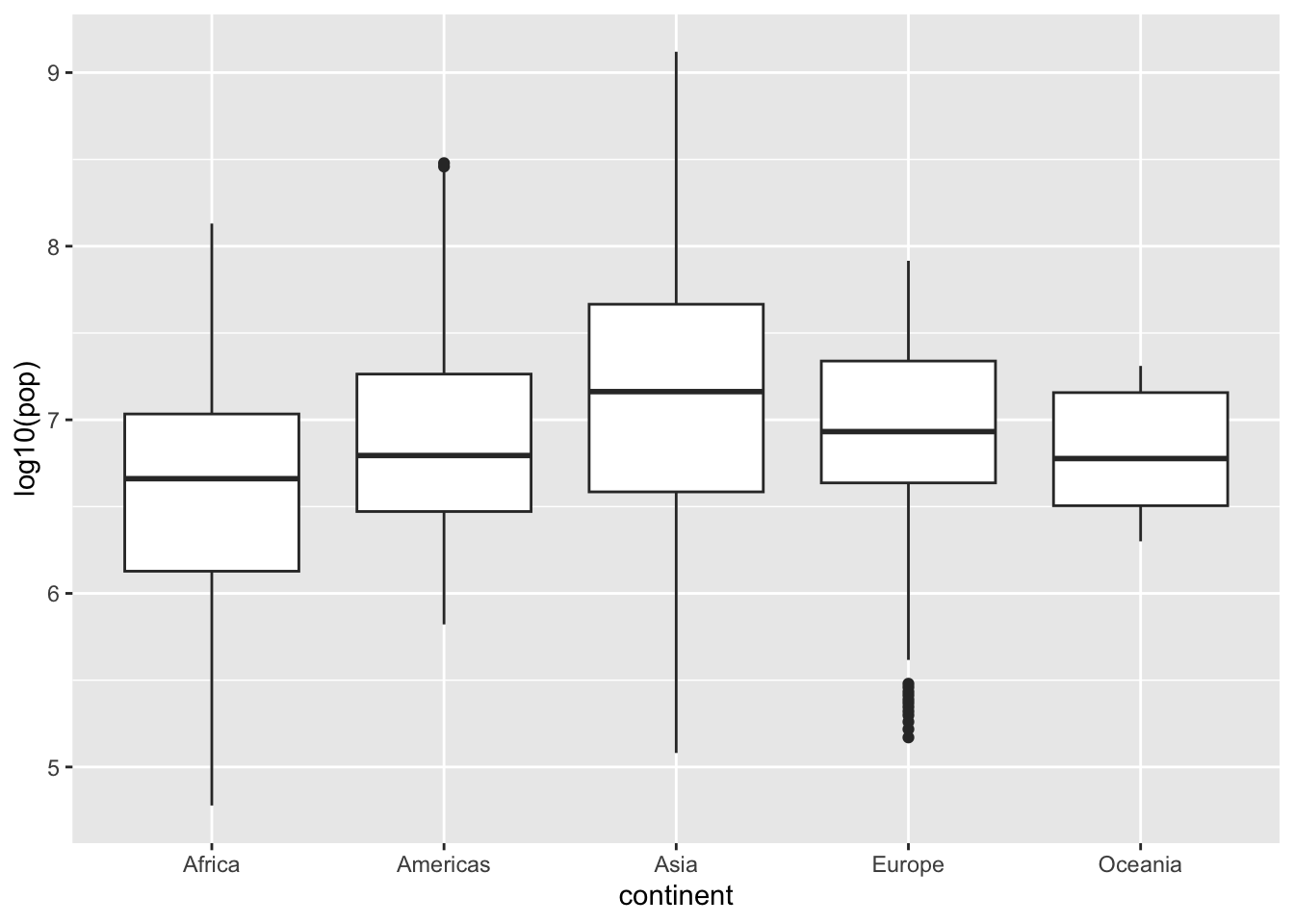
Alternately, you can use coord_trans(x = "identity", y = "log10") in stead of y = log10(pop).
Can you see the difference?
ggplot(df, aes(x = continent, y = pop)) + geom_boxplot() +
coord_trans(x = "identity", y = "log10")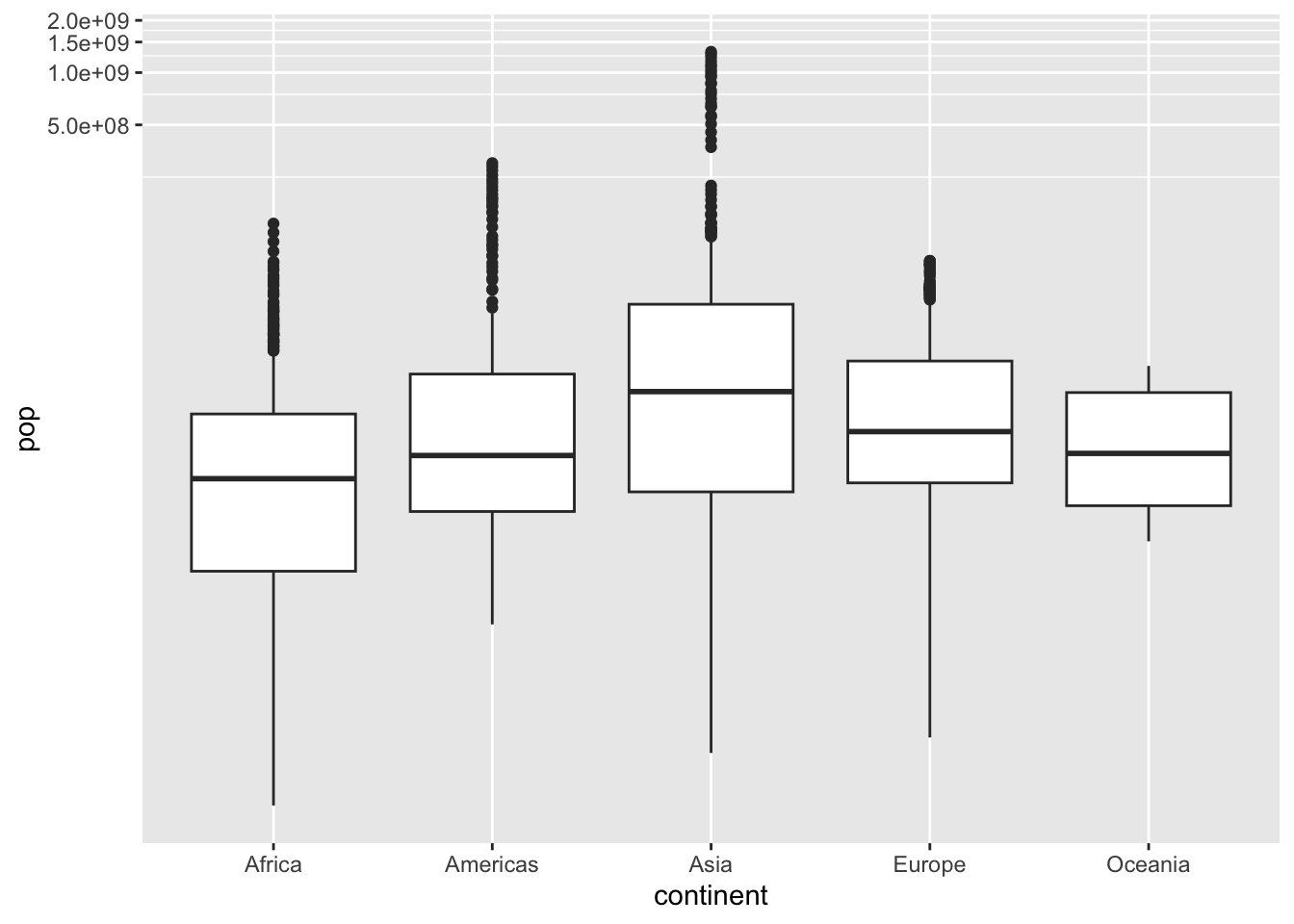
df %>% filter(year %in% c(1957, 1982, 2007)) %>%
ggplot() + geom_boxplot(aes(x = continent, y = pop, fill = factor(year))) +
coord_trans(x = "identity", y = "log10")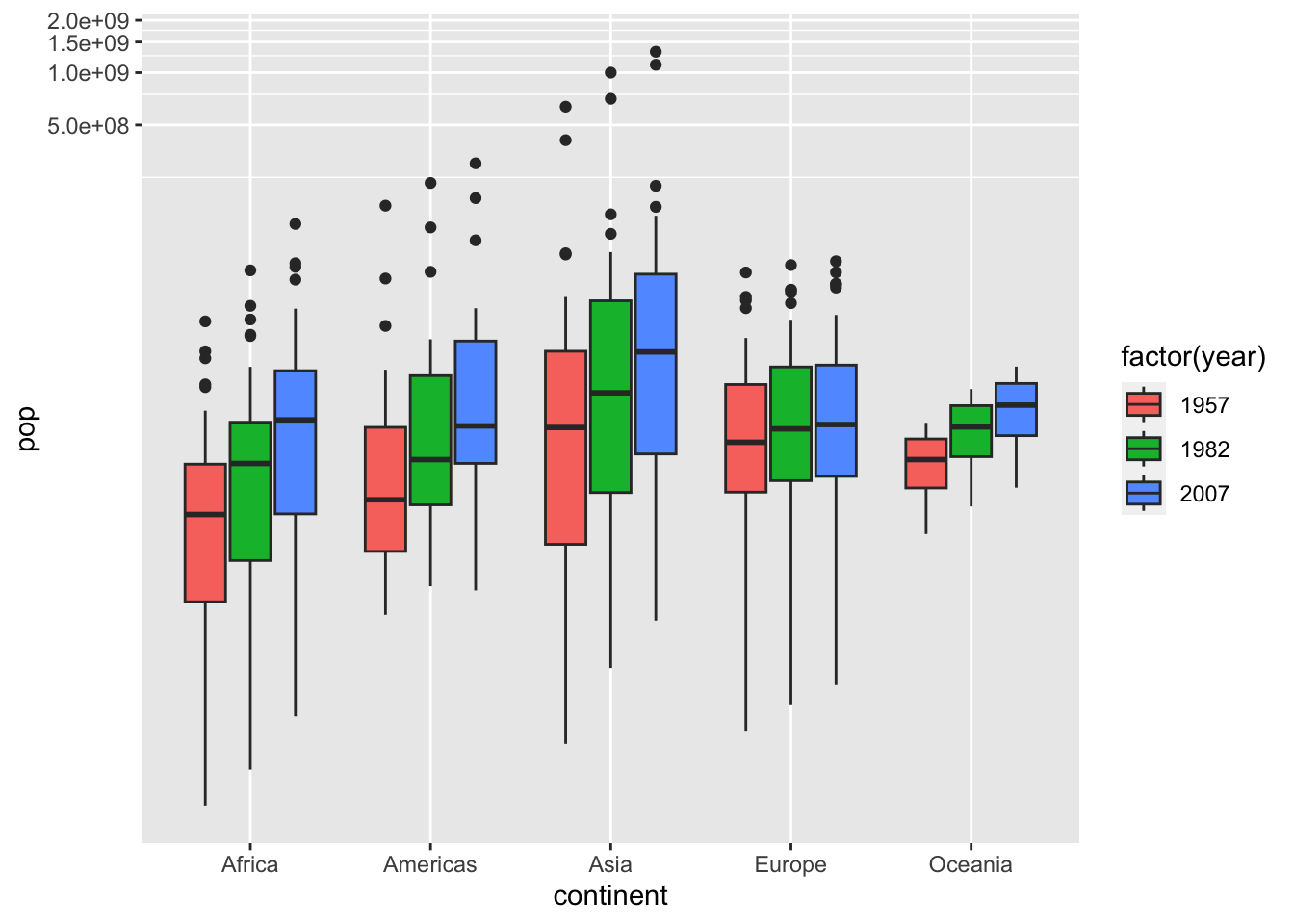
df %>% filter(year %in% c(1957, 1982, 2007)) %>%
ggplot() + geom_boxplot(aes(x = continent, y = gdpPercap, fill = factor(year))) +
coord_trans(x = "identity", y = "log10")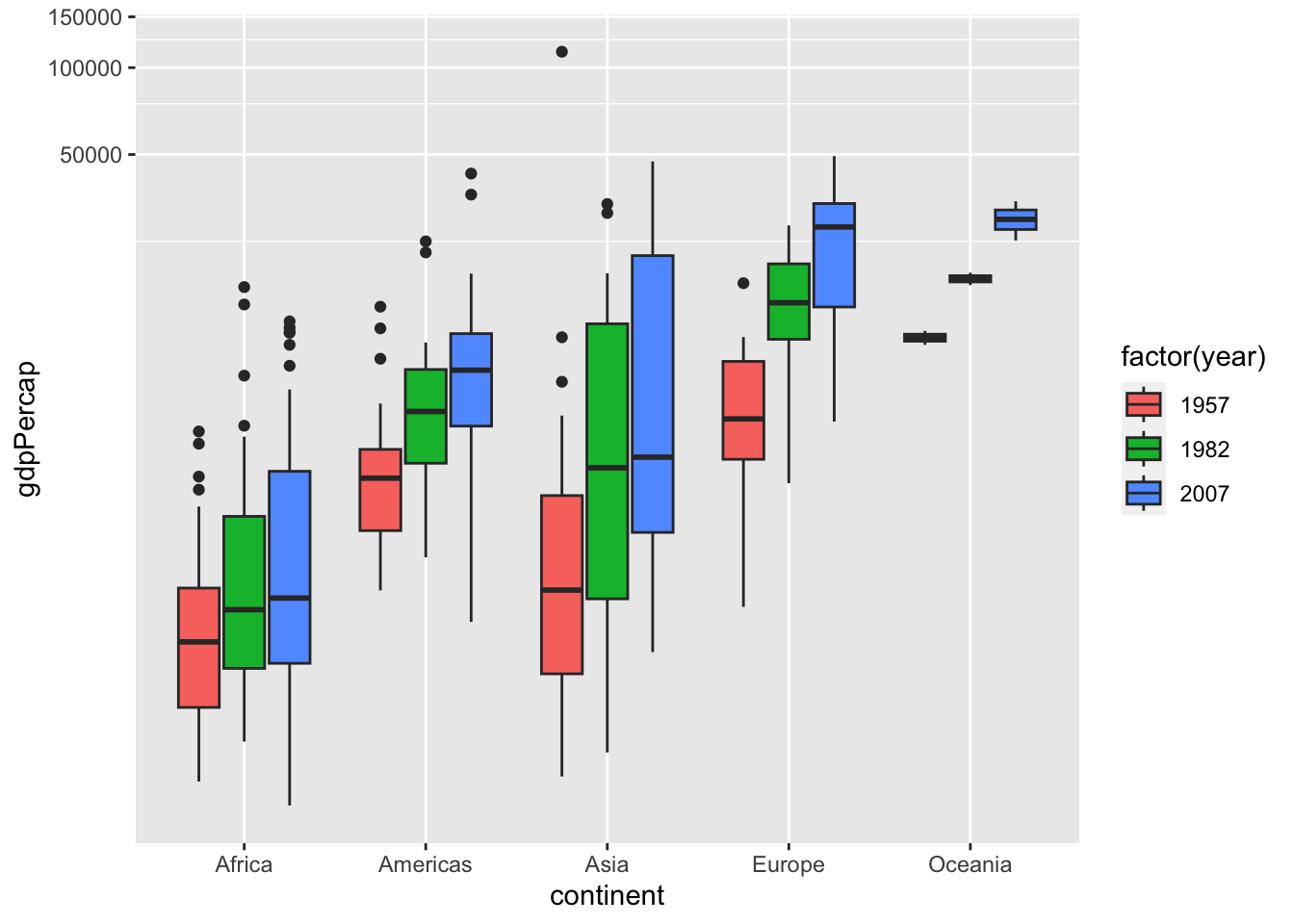
ggplot(df, aes(gdpPercap, lifeExp)) + geom_point(aes(color=continent))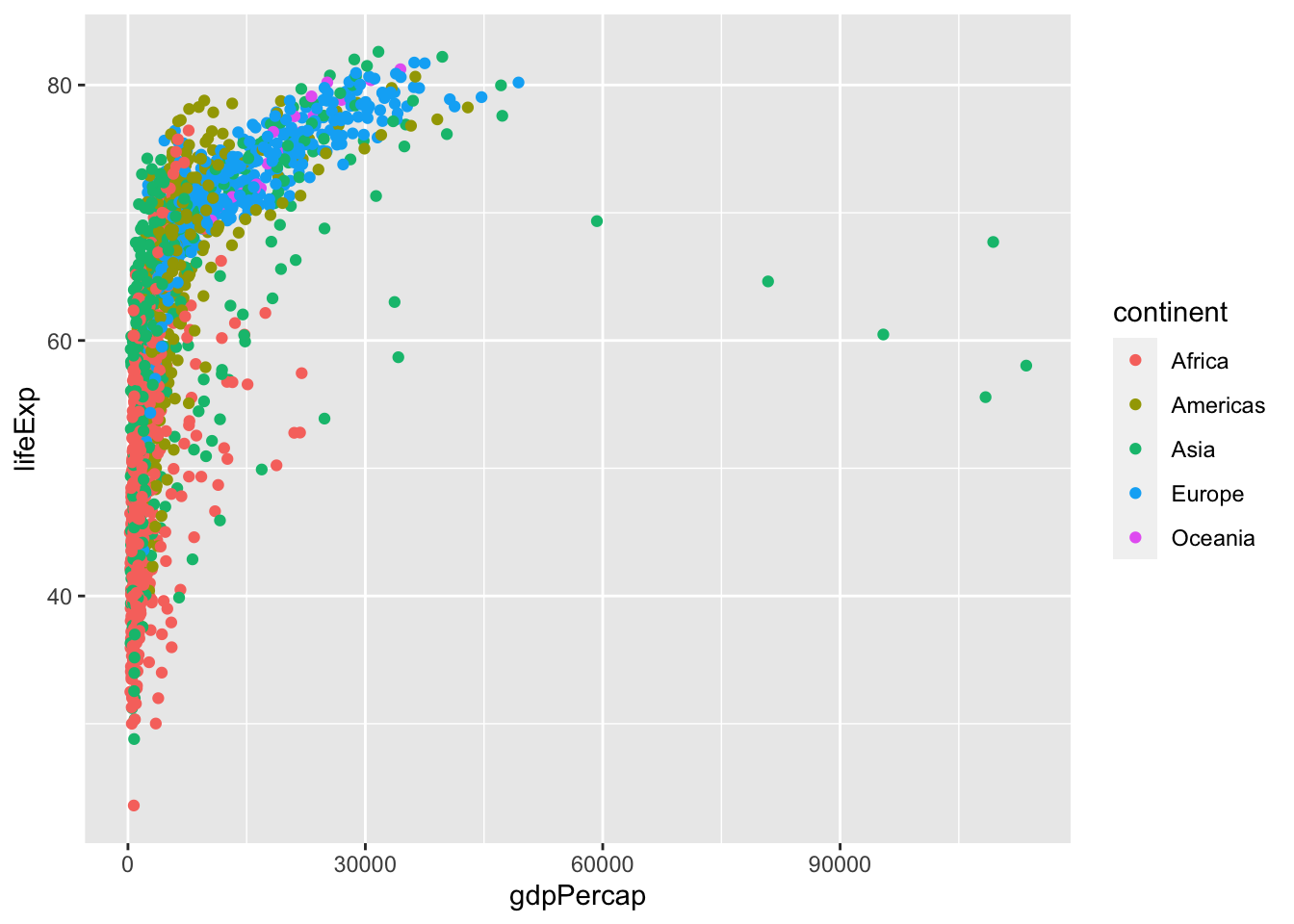
df %>% ggplot(aes(gdpPercap, lifeExp, color = continent)) + geom_point() +
coord_trans(x = "log10", y = "identity")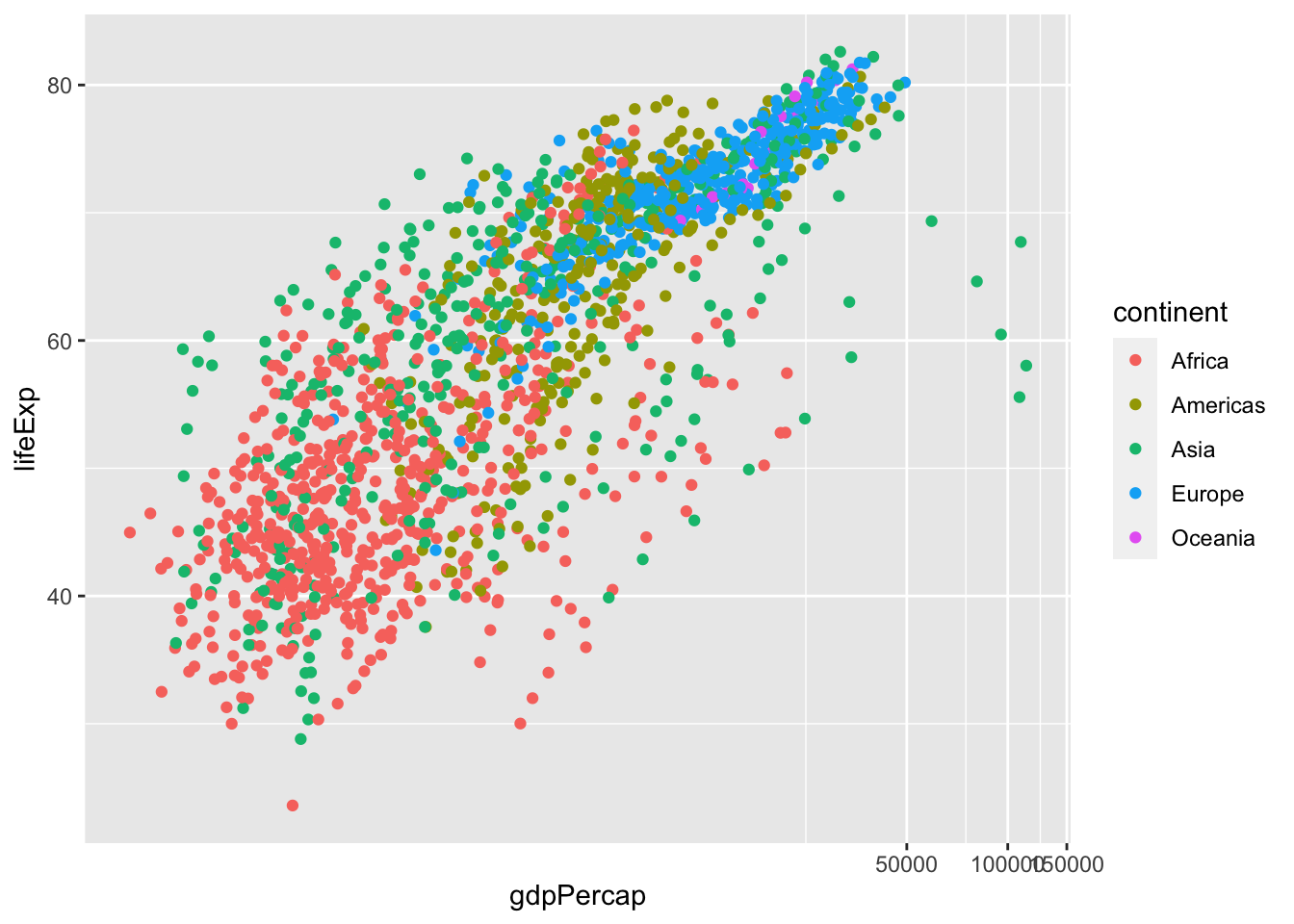
df %>% filter(year %in% c(2007)) %>%
ggplot() + geom_point(aes(x = pop, y = gdpPercap, col = continent)) +
coord_trans(x = "log10", y = "log10")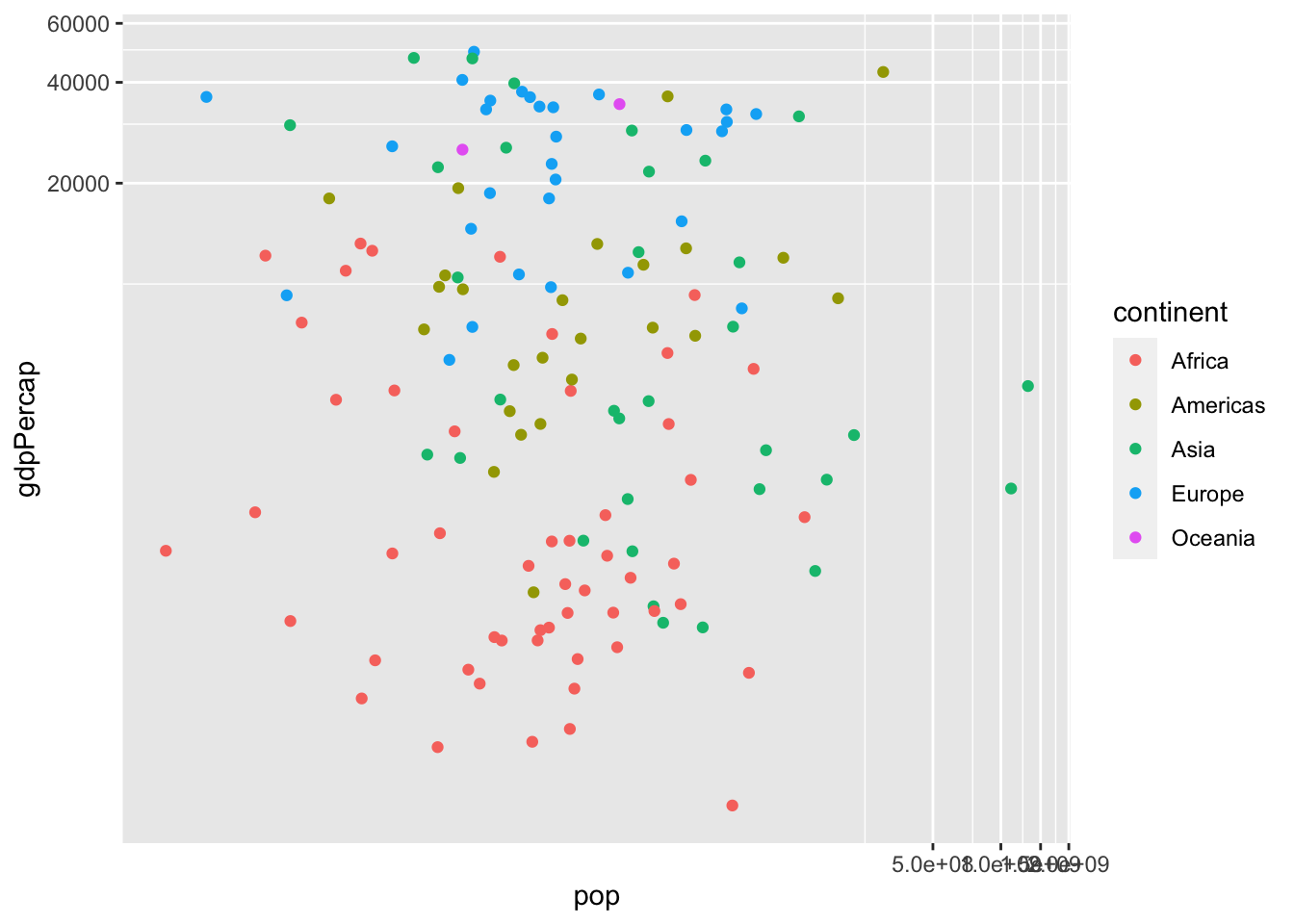
df %>% filter(year %in% c(1957)) %>%
ggplot() + geom_point(aes(x = gdpPercap, y = lifeExp, col = continent, size = pop)) +
coord_trans(x = "log10", y = "identity")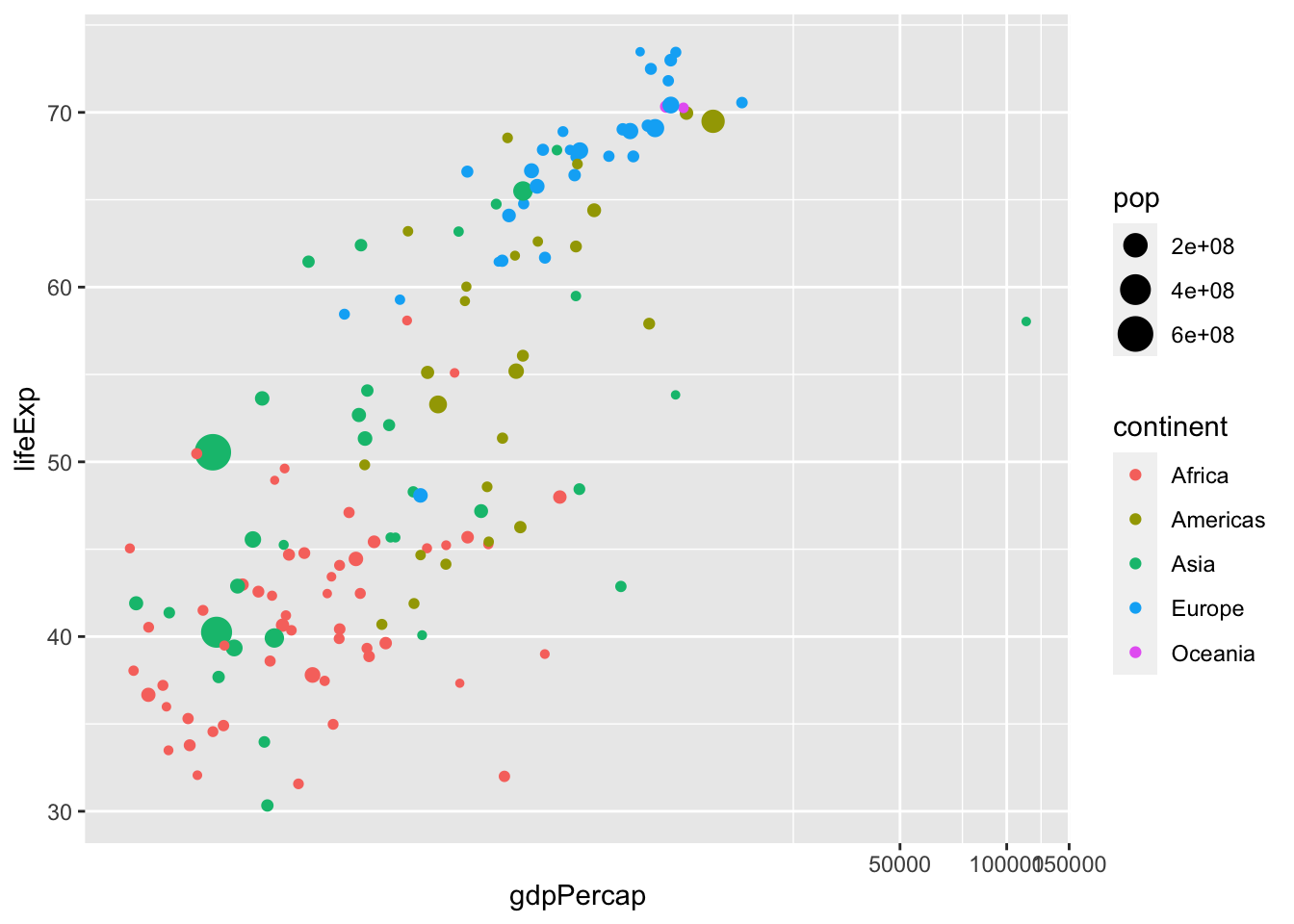
df %>% filter(year %in% c(2007)) %>%
ggplot() + geom_point(aes(x = gdpPercap, y = lifeExp, col = continent, size = pop)) +
coord_trans(x = "log10", y = "identity")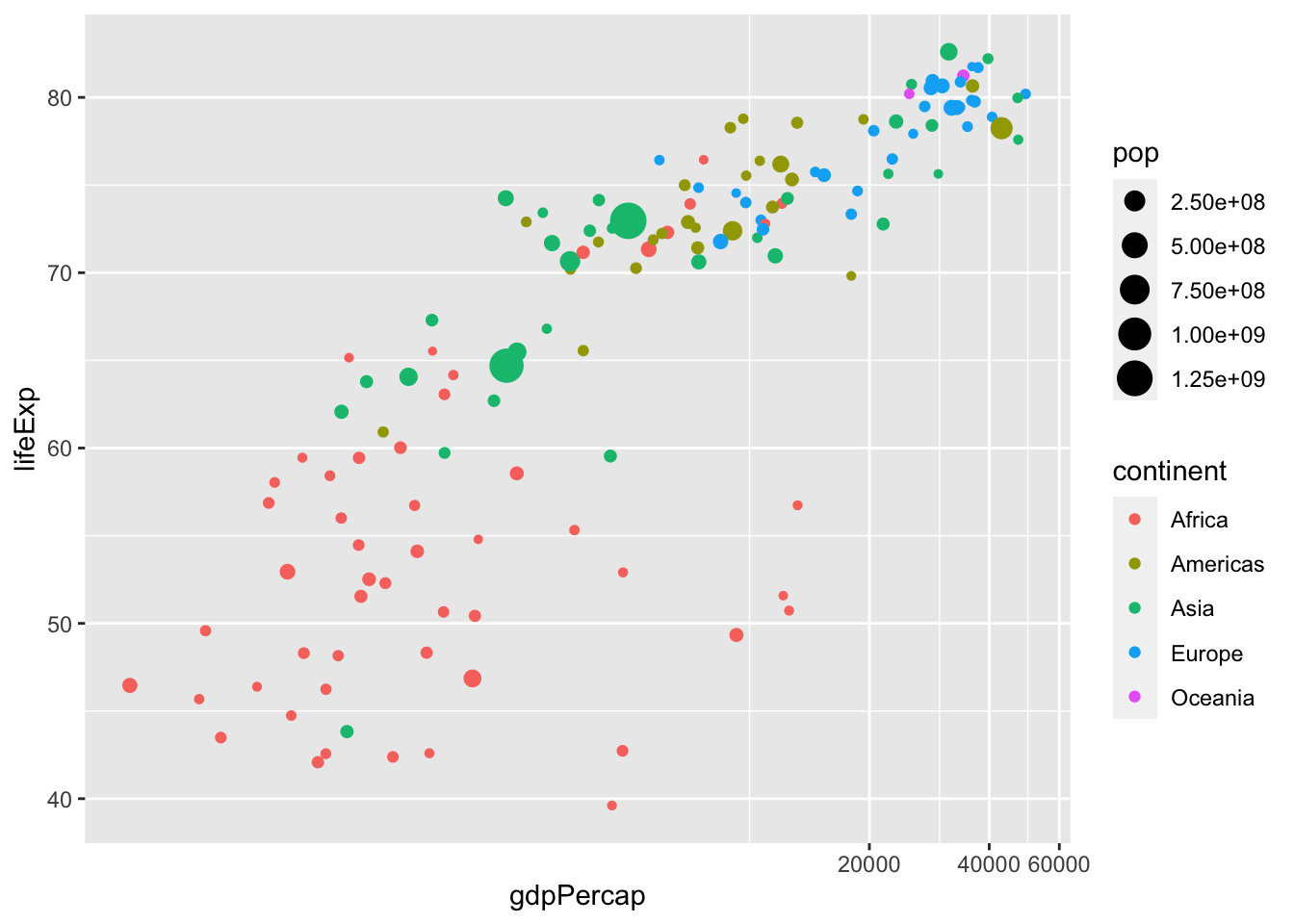
df %>% filter(year %in% c(1957, 2007)) %>%
ggplot() + geom_point(aes(x = gdpPercap, y = lifeExp, col = continent, size = pop)) +
coord_trans(x = "log10", y = "identity") +
facet_wrap(vars(year))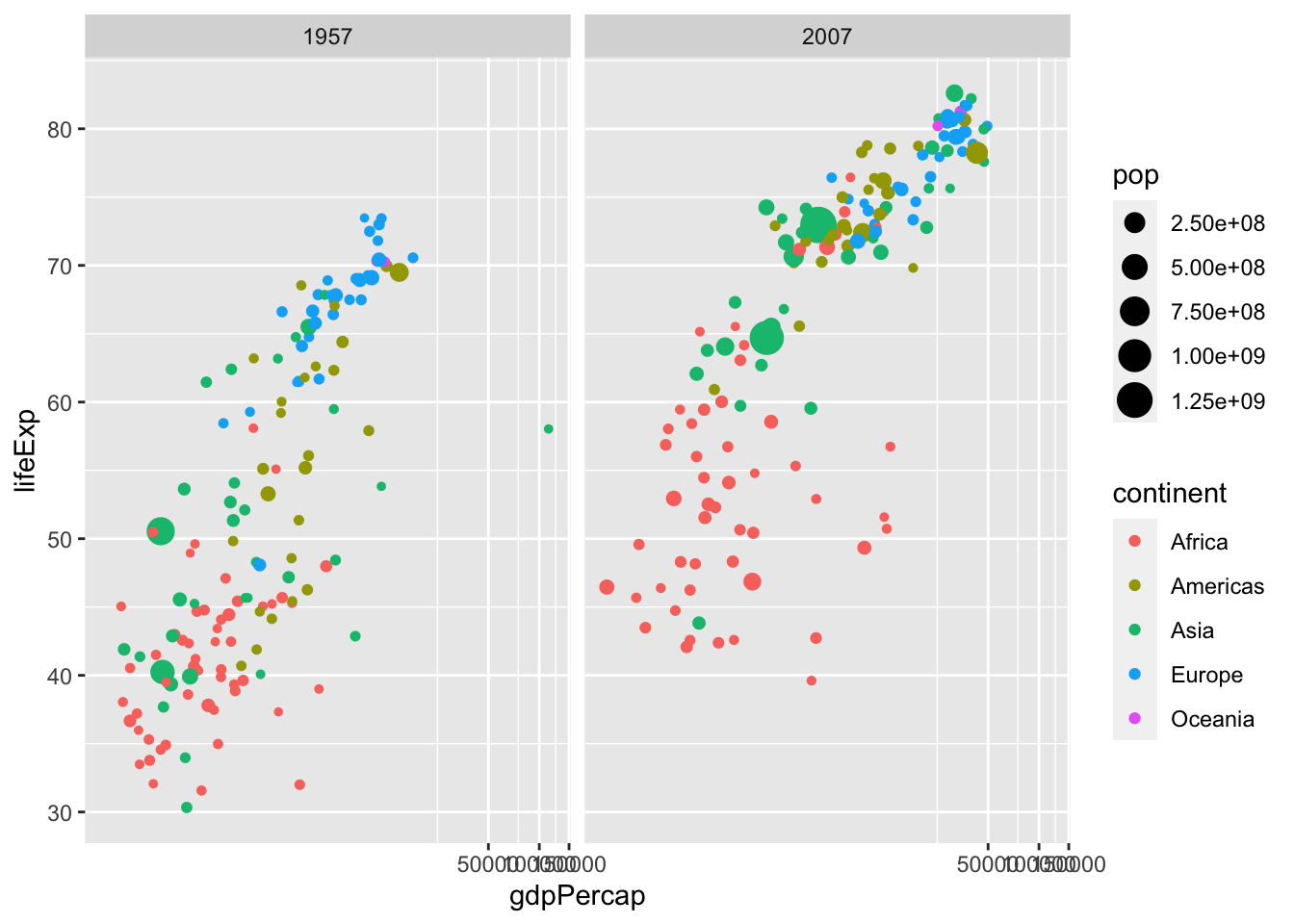
df_lifeExp <- df %>% group_by(continent, year) %>%
summarize(mean_lifeExp = mean(lifeExp), median_lifeExp = median(lifeExp), max_lifeExp = max(lifeExp), min_lifeExp = min(lifeExp))
#> `summarise()` has grouped output by 'continent'. You can
#> override using the `.groups` argument.The code above gives a message, but it works.
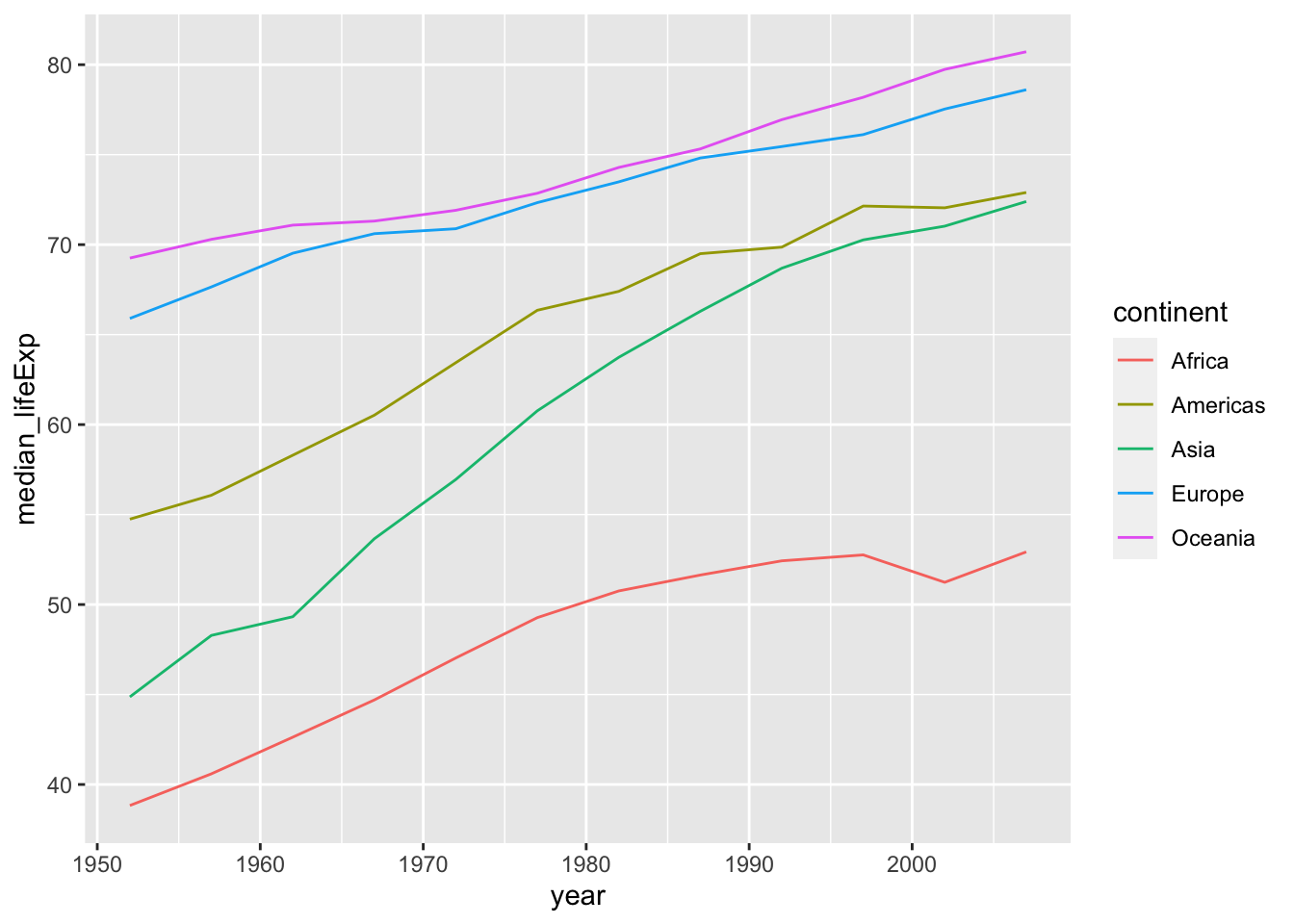
If you do not want to have a message, the following is an option. Otherwise, grouping is kept and you can get the original data back by ungroup().
df_pop <- df %>% group_by(continent, year) %>%
summarize(mean_pop = mean(pop), median_pop = median(pop), max_pop = max(pop), min_pop = min(pop), .groups = "drop")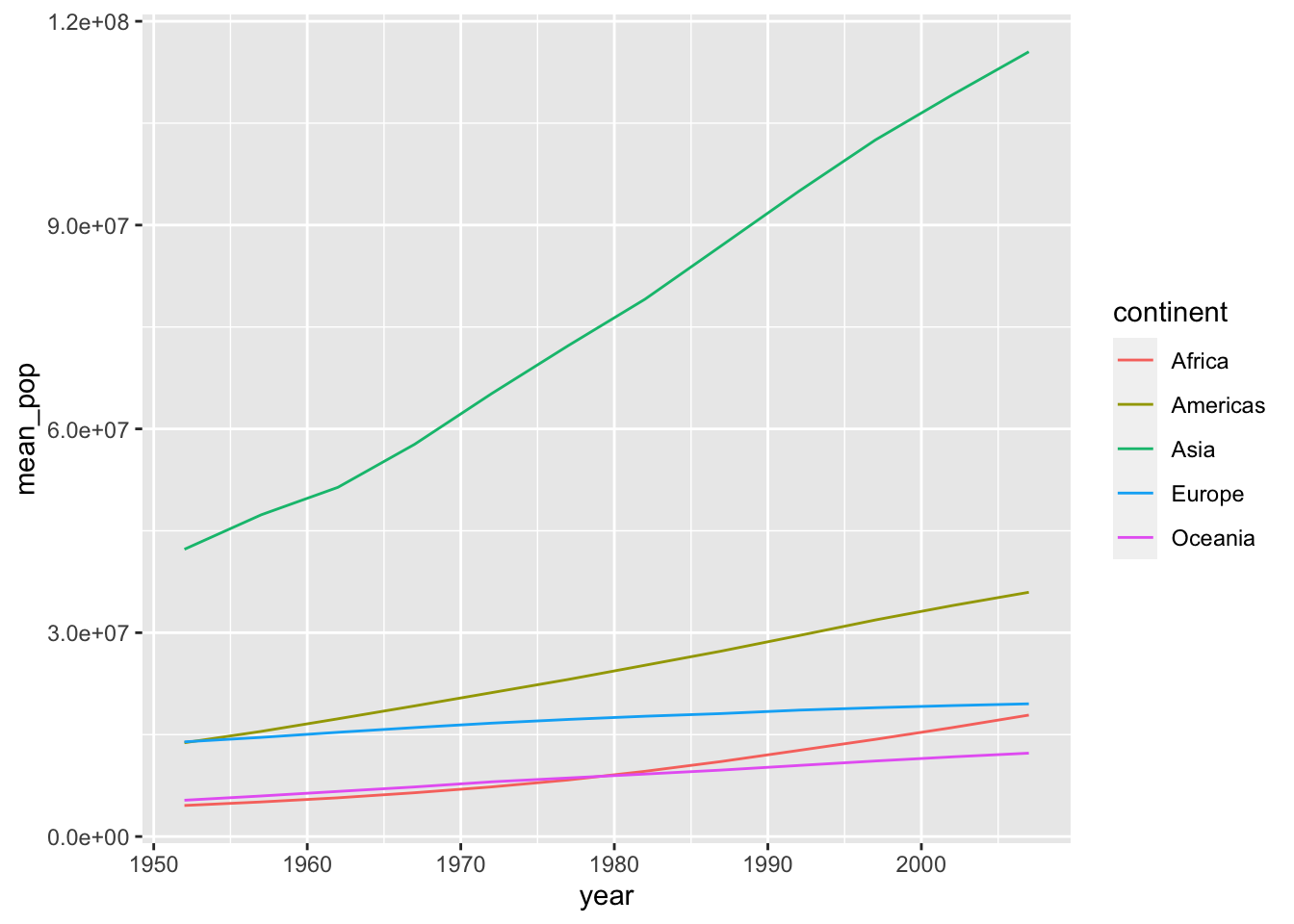 The following two codes create the same chart.
The following two codes create the same chart.
df_pop %>% ggplot(aes(x = year, y = mean_pop, color = continent, linetype = continent)) +
geom_line()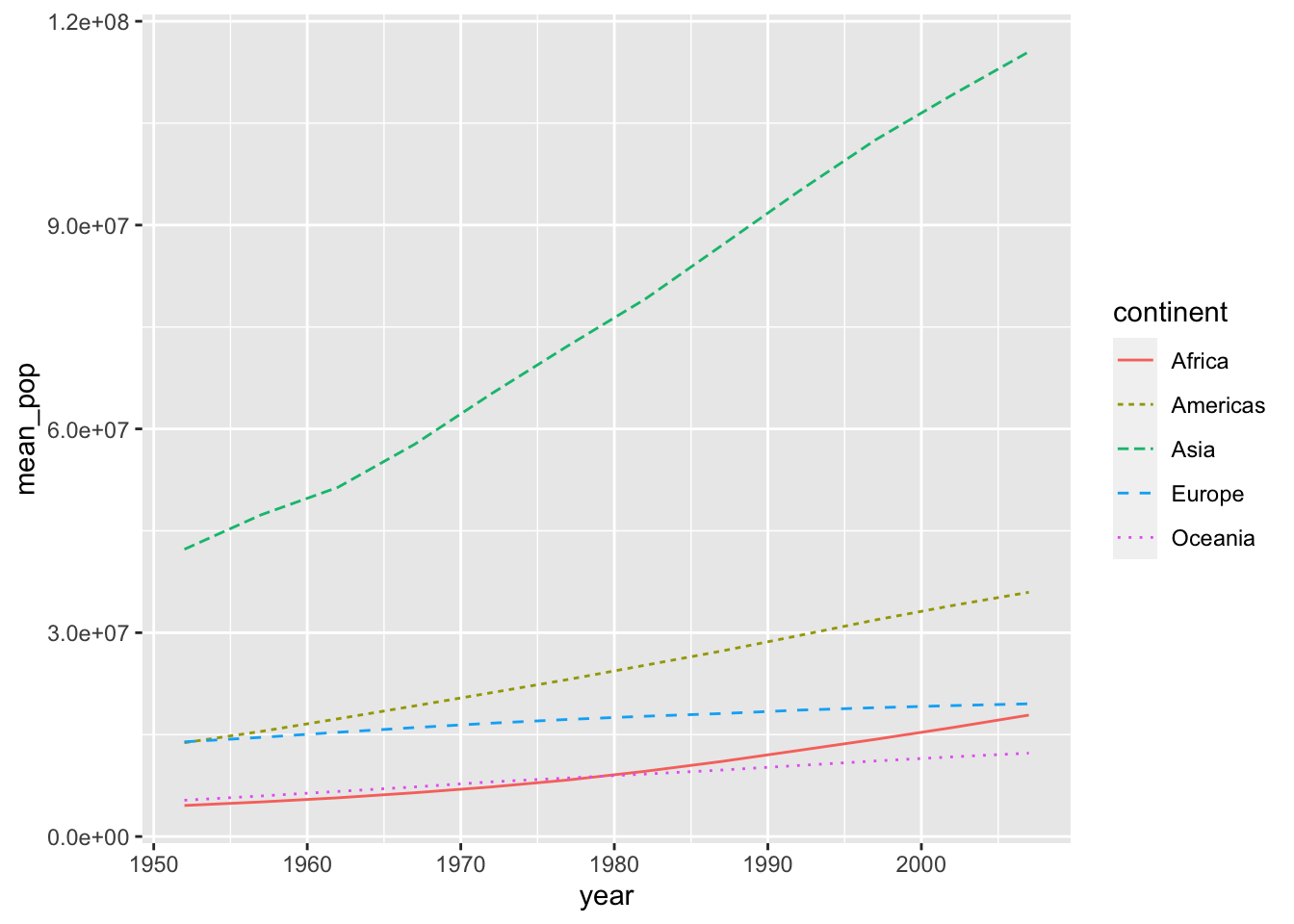
df_pop %>% ggplot() +
geom_line(aes(x = year, y = mean_pop, color = continent)) +
geom_line(aes(x = year, y = median_pop, linetype = continent))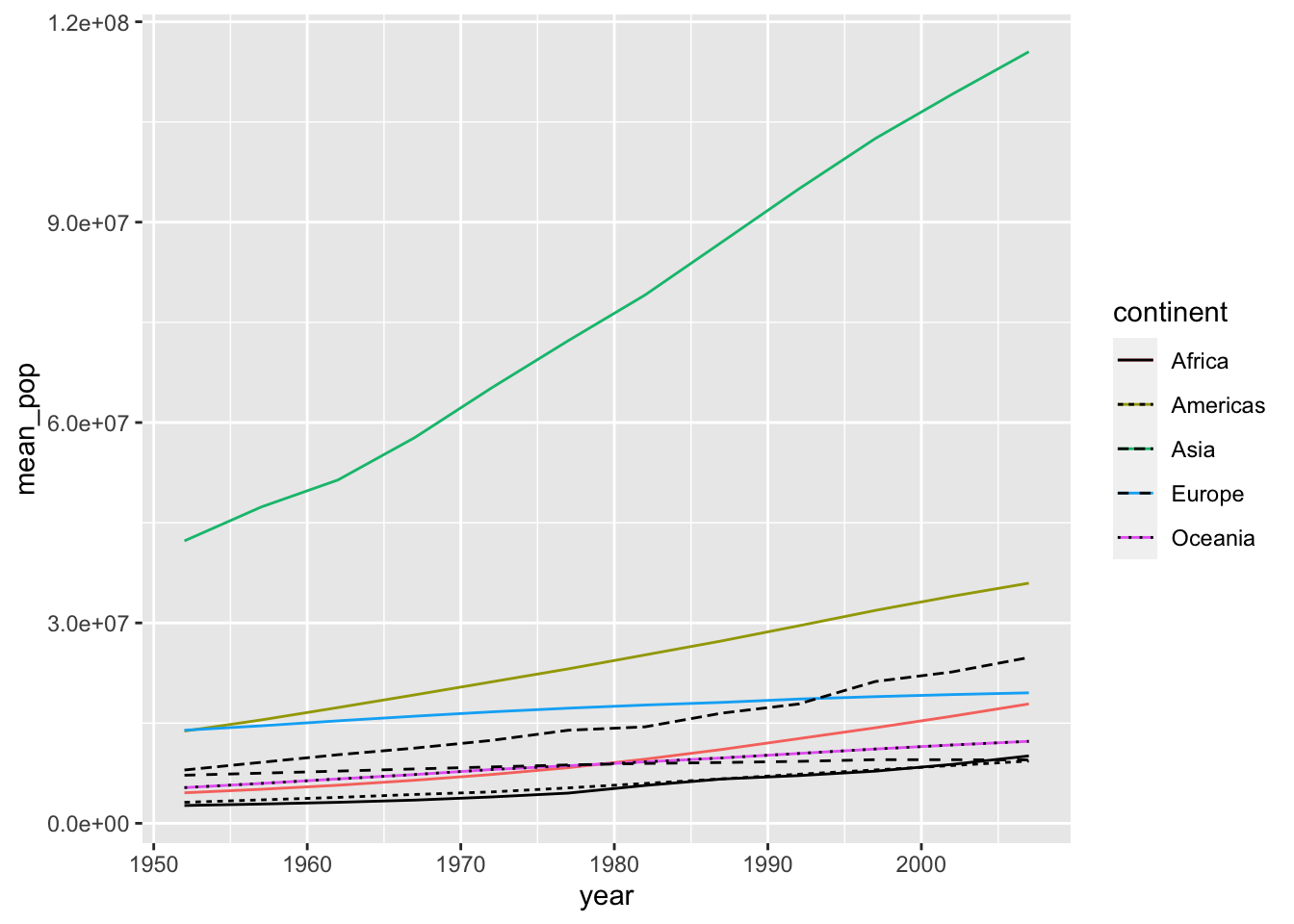
6.3 Responses to Questions
6.3.1 Q1. Two categorical variables and one numerical variables
Eg. Smoking, Alcohol and (O)esophageal Cancer
(df_esoph <- as_tibble(esoph))
#> # A tibble: 88 × 5
#> agegp alcgp tobgp ncases ncontrols
#> <ord> <ord> <ord> <dbl> <dbl>
#> 1 25-34 0-39g/day 0-9g/day 0 40
#> 2 25-34 0-39g/day 10-19 0 10
#> 3 25-34 0-39g/day 20-29 0 6
#> 4 25-34 0-39g/day 30+ 0 5
#> 5 25-34 40-79 0-9g/day 0 27
#> 6 25-34 40-79 10-19 0 7
#> 7 25-34 40-79 20-29 0 4
#> 8 25-34 40-79 30+ 0 7
#> 9 25-34 80-119 0-9g/day 0 2
#> 10 25-34 80-119 10-19 0 1
#> # ℹ 78 more rowsdf_esoph has three categorical variables and one numerical variable ncases to investigate.
Comments: I wanted to include three variables in the first exercise to be able to compare tobacco consumption, number of cases of cancer, and age in the same graph but I was not able to do it.
Solutions: There are various ways you can choose from.
Scatter plot with size by geom_point().
ggplot(df_esoph) + geom_point(aes(agegp, tobgp, size=ncases))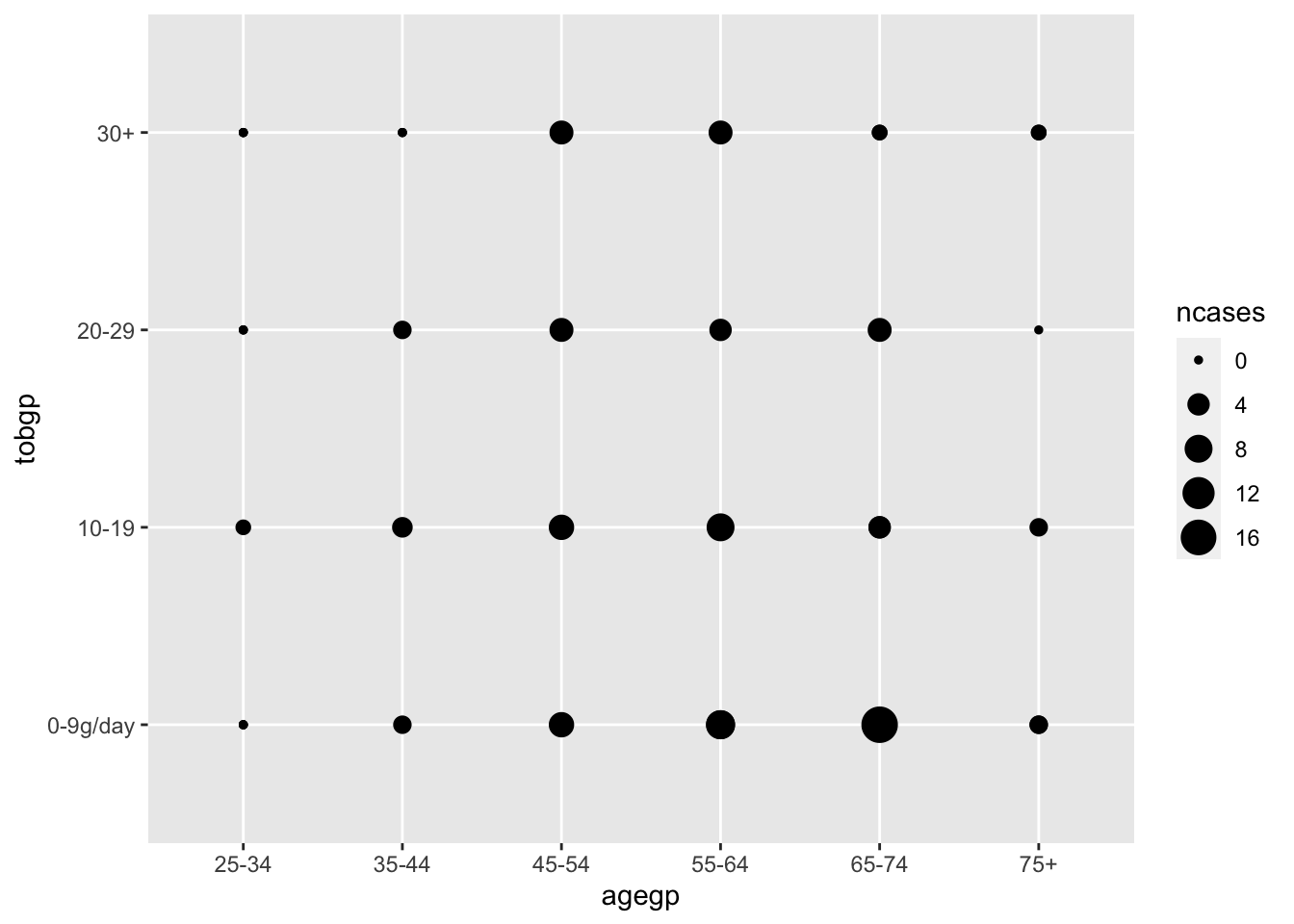
Heatmap with geom_tile()
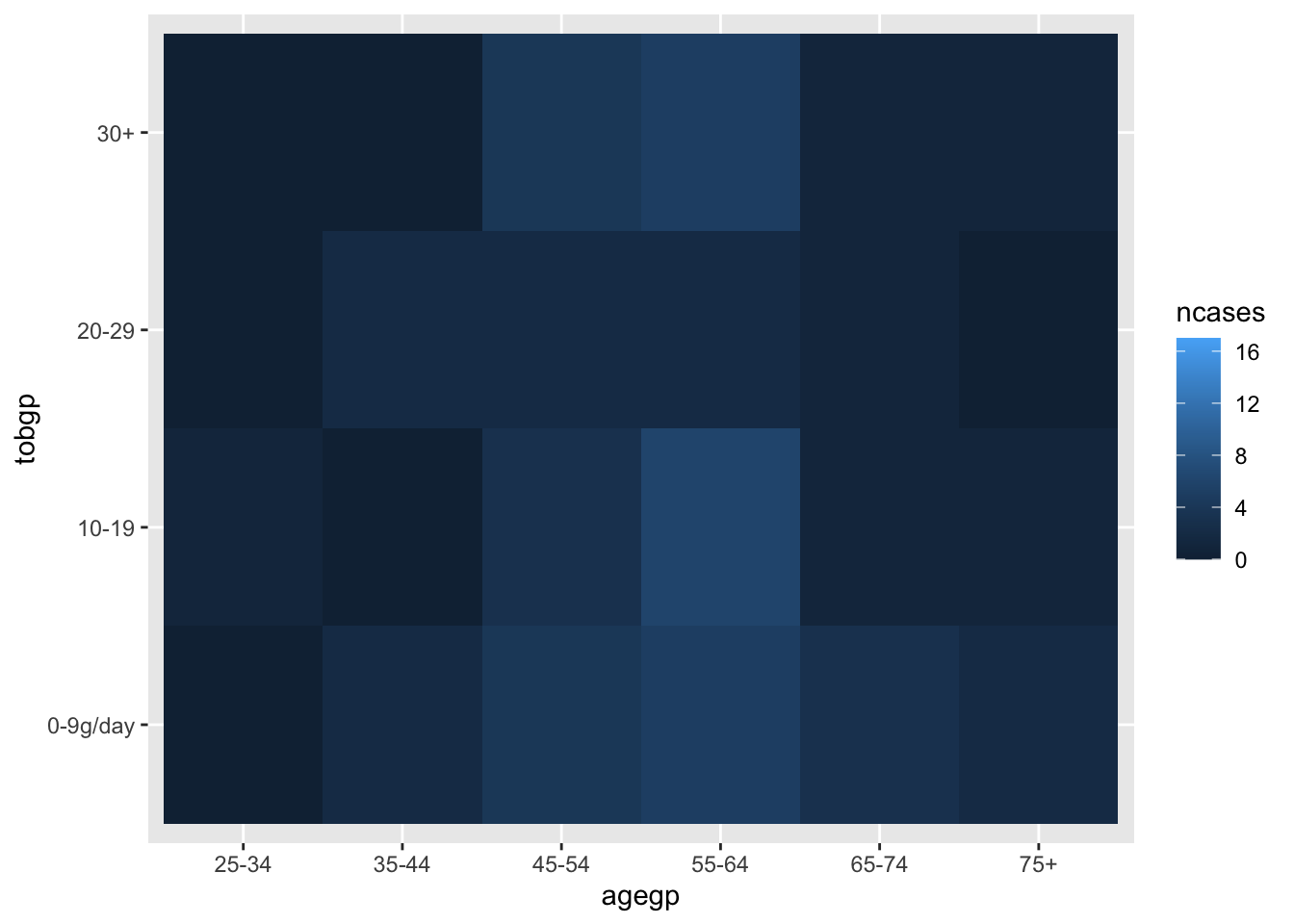
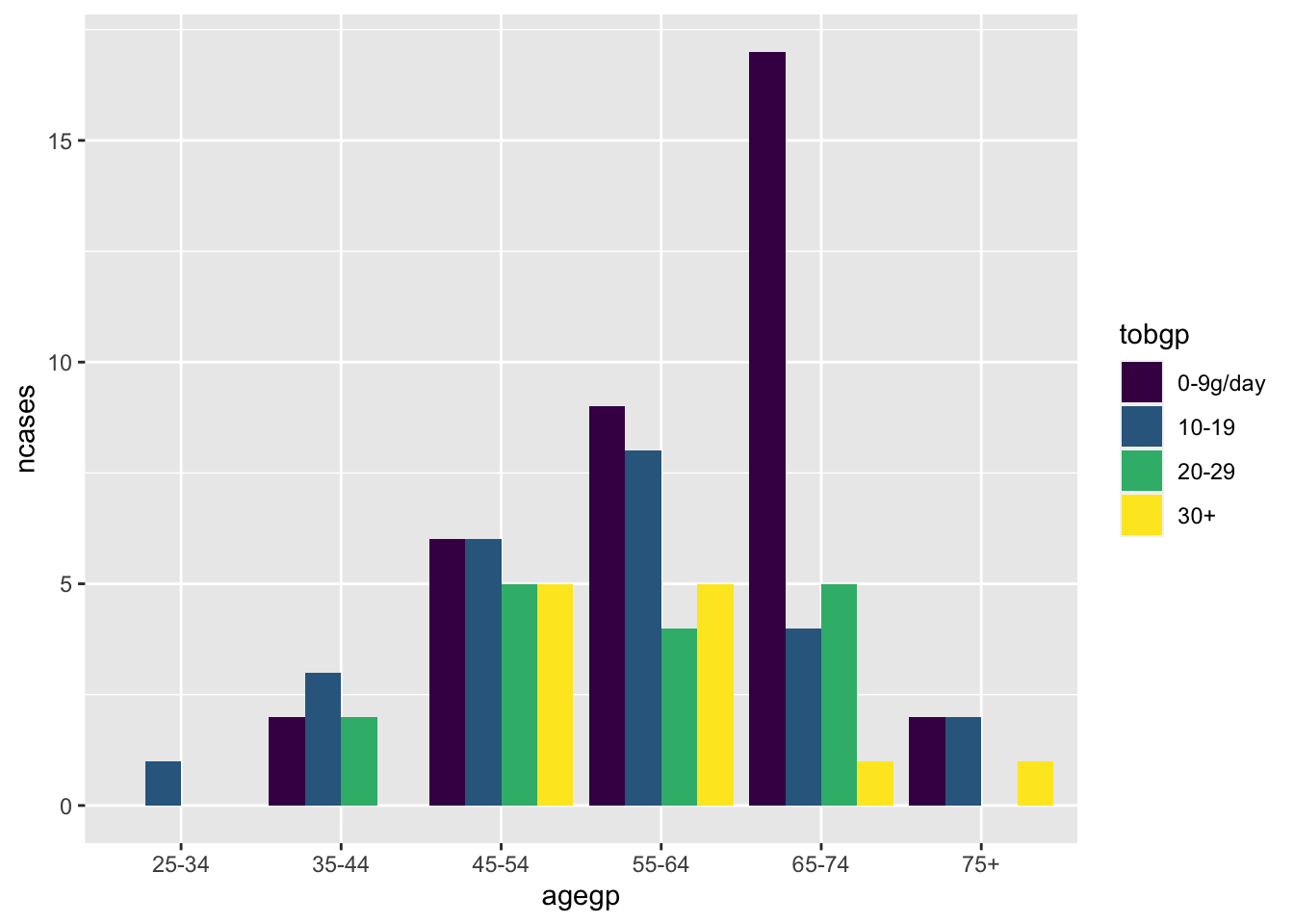
ggplot(df_esoph, aes(x= tobgp, y=ncases, fill=agegp))+geom_boxplot()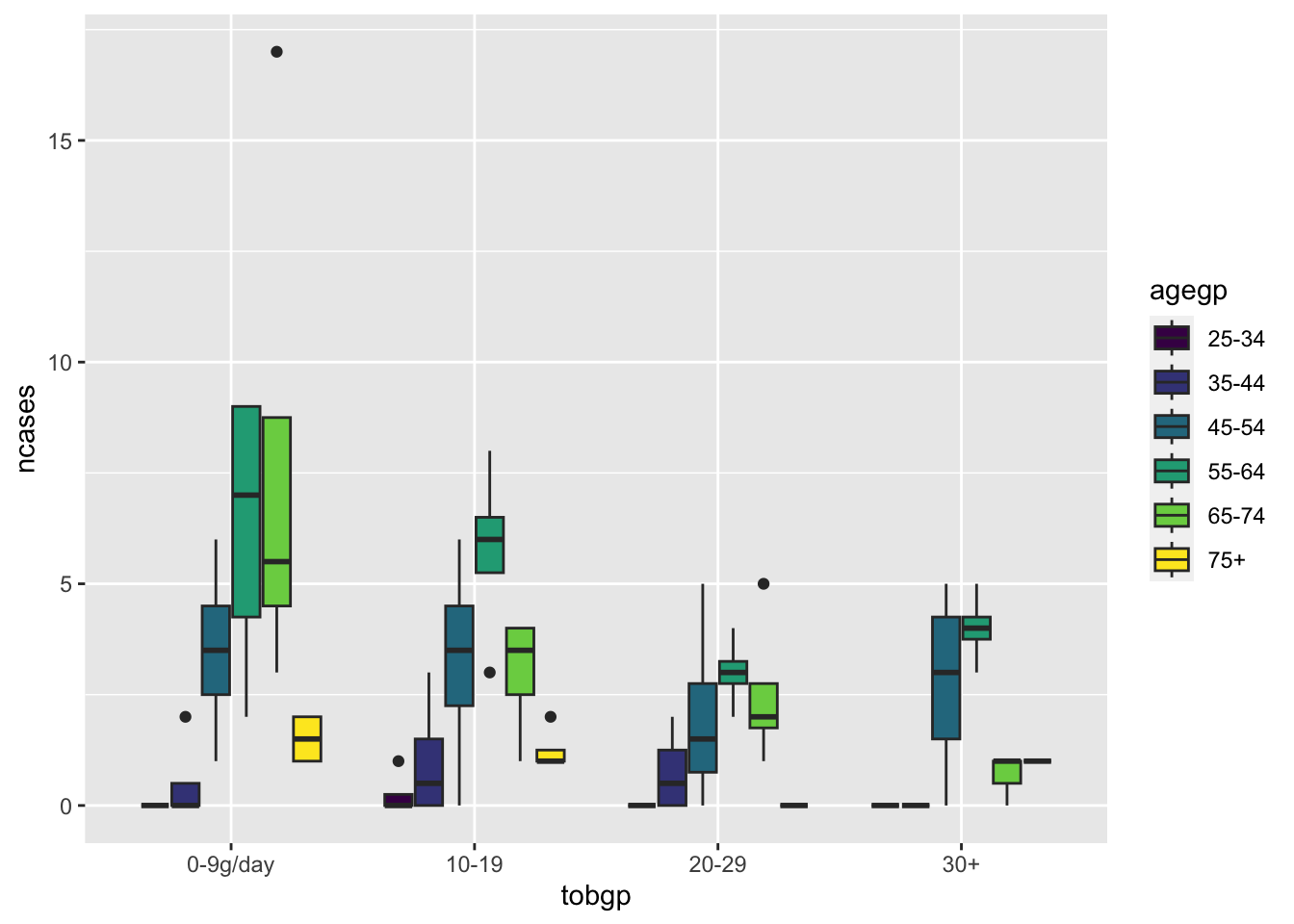
ggplot(df_esoph, aes(x= agegp, y=ncases, fill=tobgp))+geom_boxplot()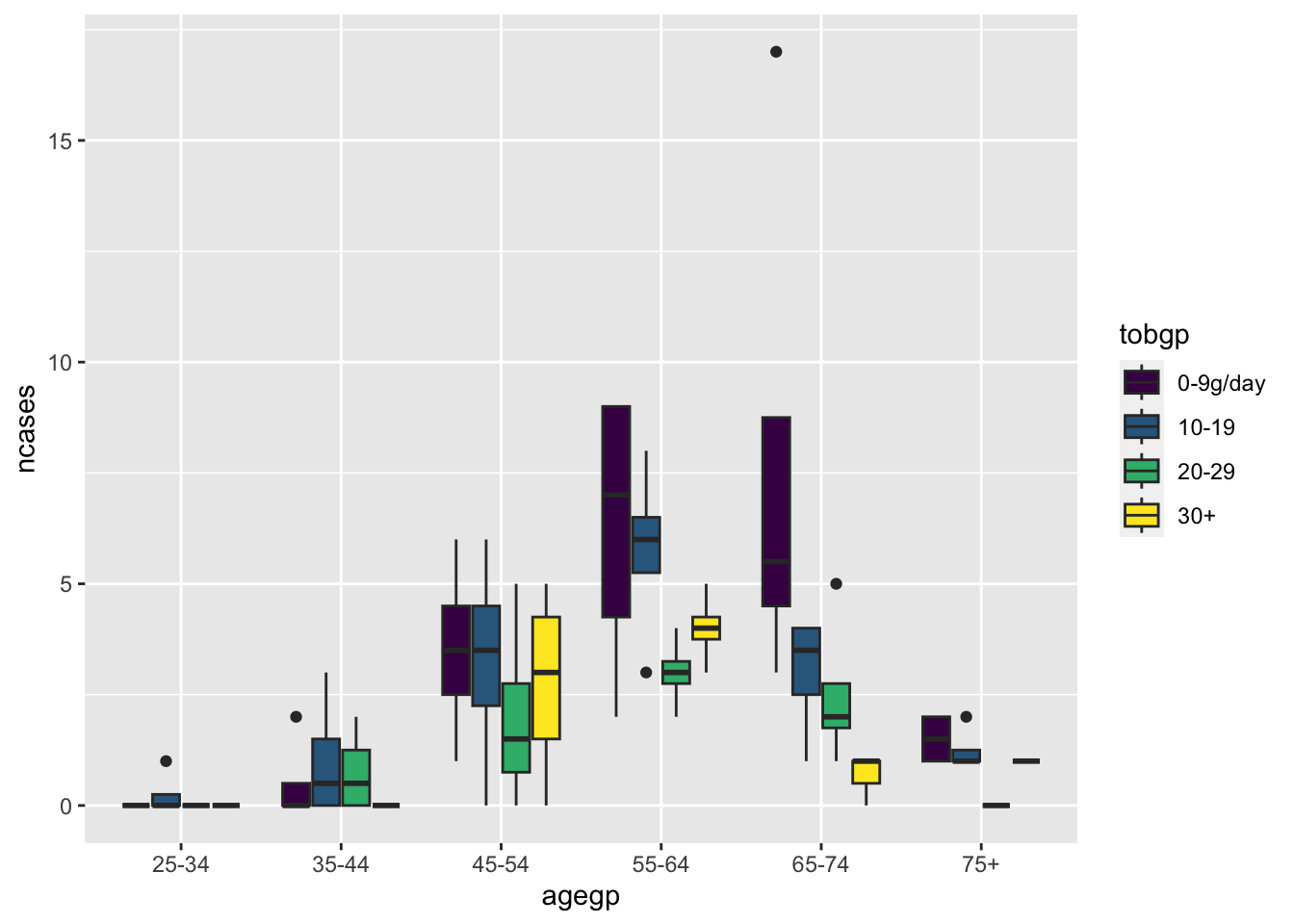
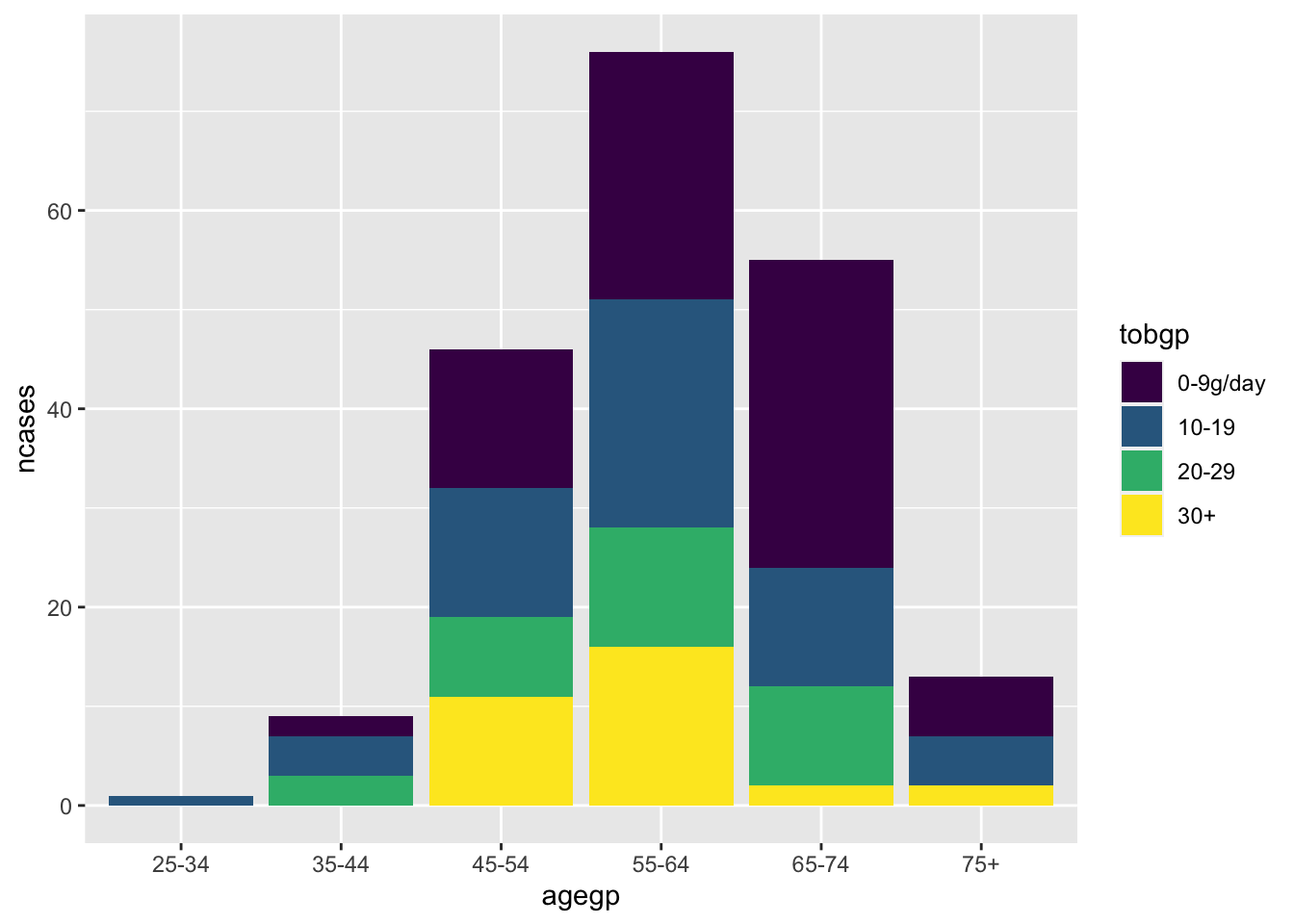
Default position is “stack”.
6.3.2 Q2. Combine two charts
df %>%
select(country, continent, year, gdpPercap) %>%
filter(continent %in% c("Asia", "Europe")) %>%
group_by(continent, year) %>%
summarise(mean_GDPperCapita = mean(gdpPercap)) %>%
ggplot(aes(x=year)) +
geom_line(aes(y=mean_GDPperCapita, color=continent)) +
ggtitle("GDP oer capita by continents, 1950's to today")
#> `summarise()` has grouped output by 'continent'. You can
#> override using the `.groups` argument.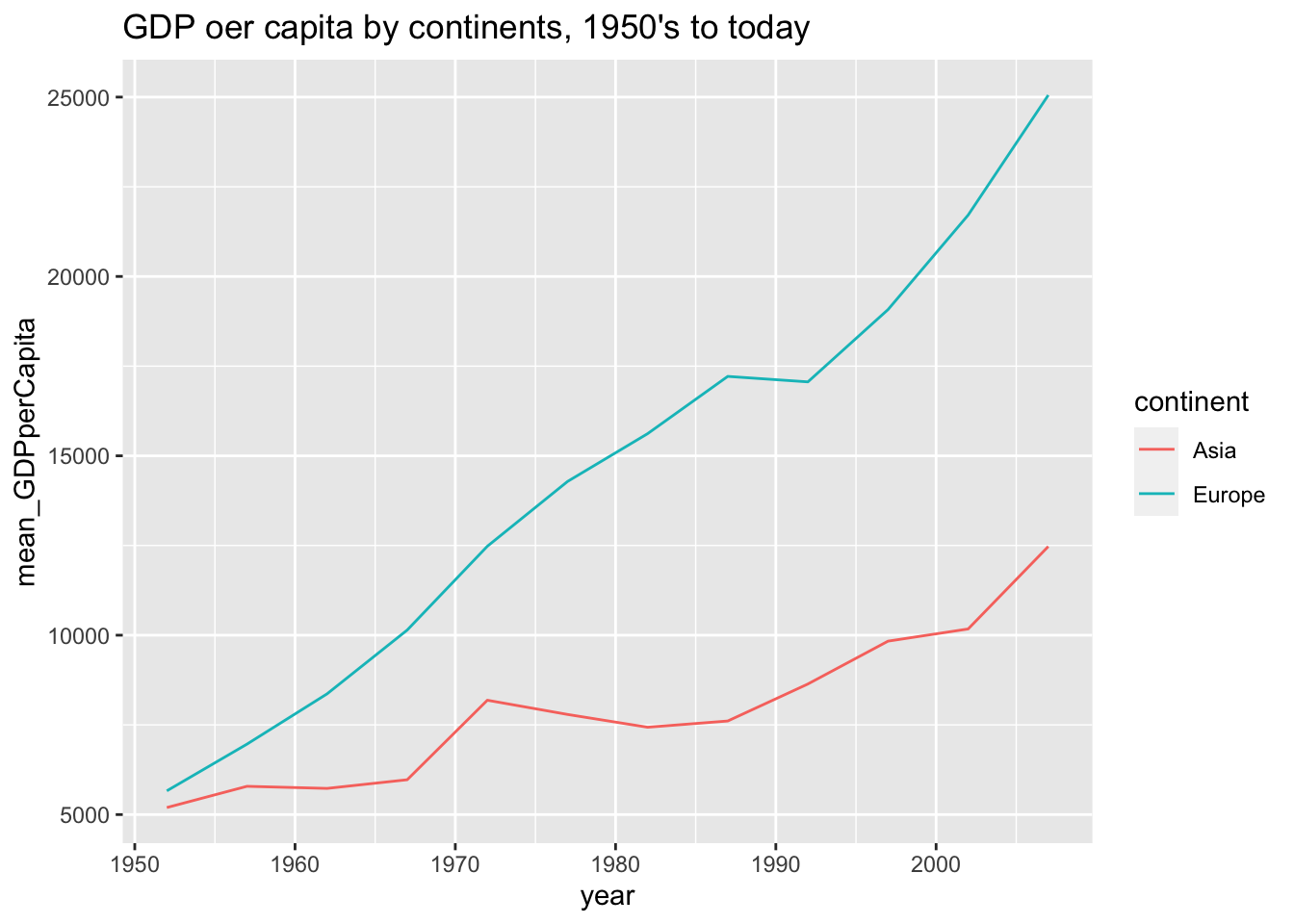
df %>%
select(country, year, gdpPercap) %>%
filter(country %in% c("Israel", "Japan", "Norway", "China", "Ireland")) %>%
ggplot(aes(x=year)) +
geom_line(aes(y=gdpPercap, color=country))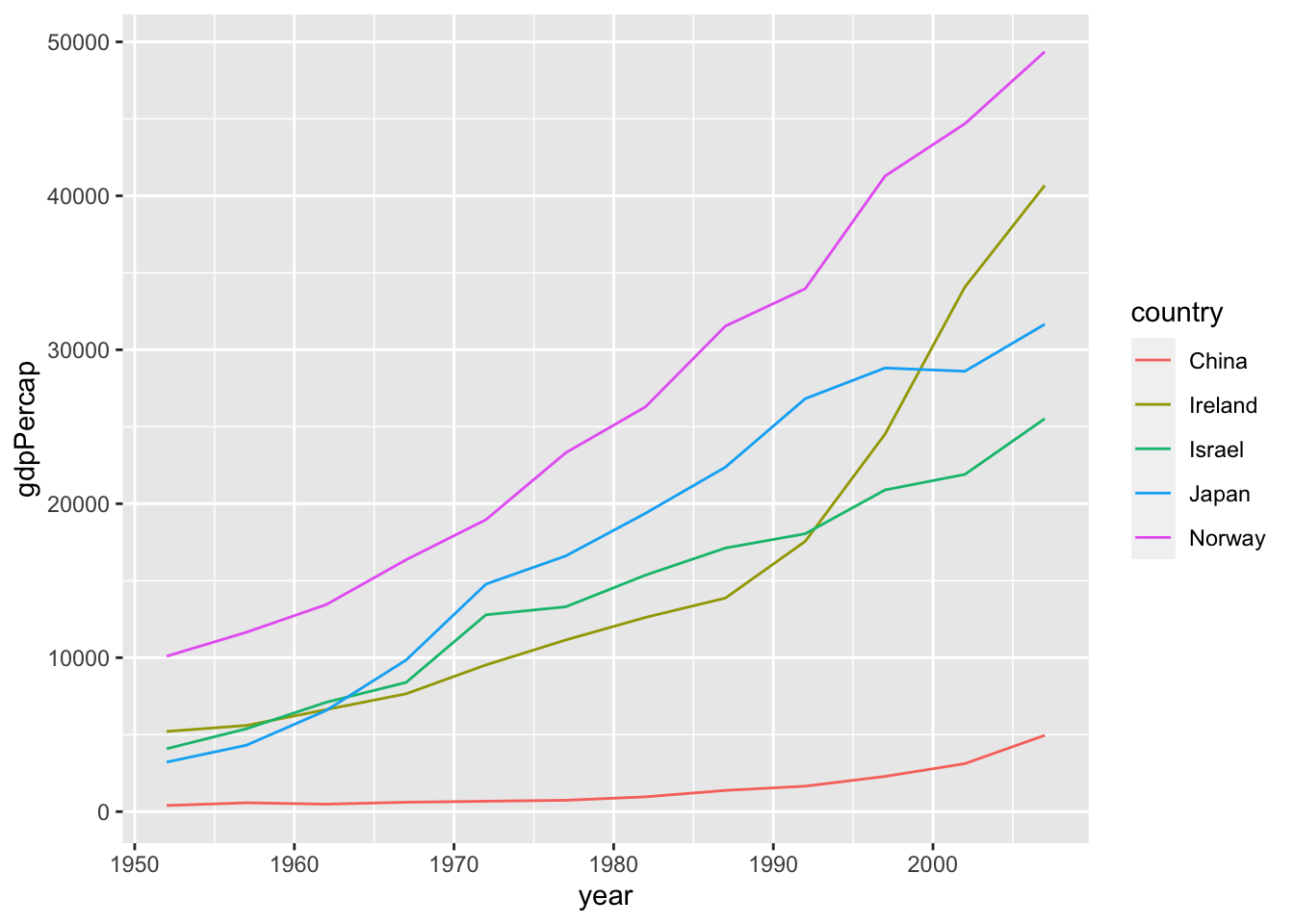
Question. I have not managed to add on the same graph of the continents the data for the individual countries, as I would have liked:
Solution. Construct two data sets and combine them into one.
ggplot2 starts with one data.
df_2c <- df %>%
select(continent, year, gdpPercap) %>%
filter(continent %in% c("Asia", "Europe")) %>%
group_by(continent, year) %>%
summarise(gdpPercap = mean(gdpPercap), .groups = 'drop') %>%
select(country = continent, year, gdpPercap)
df_5c <- df %>%
select(country, year, gdpPercap) %>%
filter(country %in% c("Israel", "Japan", "Norway", "China", "Ireland"))
df_2c %>% bind_rows(df_5c) %>%
ggplot(aes(x = year, y = gdpPercap, color = country)) + geom_line()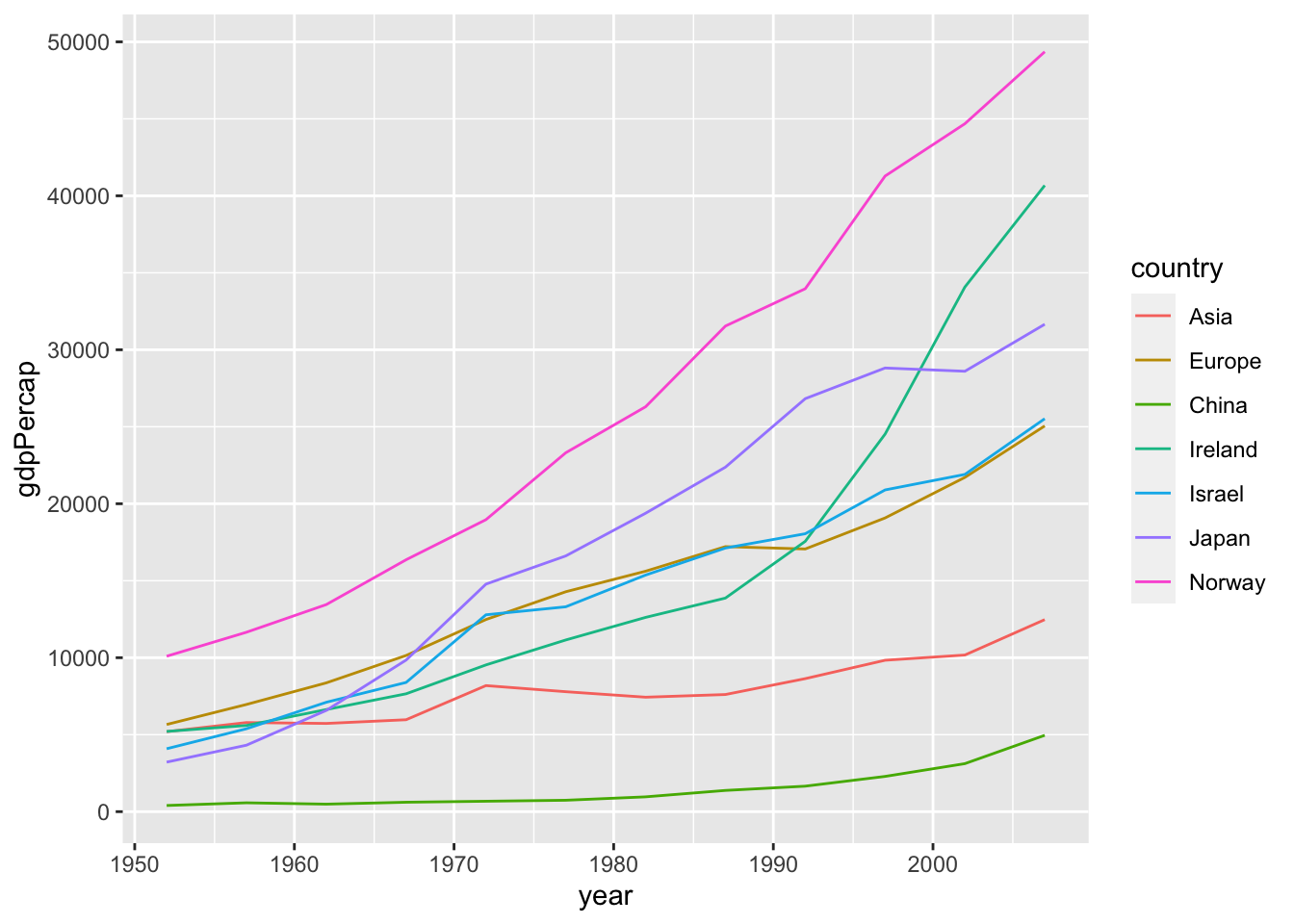
Use mutate.
df %>%
group_by(continent, year) %>%
mutate(mean_by_continent = mean(gdpPercap)) %>%
ungroup() %>%
filter(country %in% c("Israel", "Japan", "Norway", "China", "Ireland")) %>%
ggplot(aes(x = year)) +
geom_line(aes(y = gdpPercap, color=country)) +
geom_line(aes(y = mean_by_continent, linetype=continent)) +
labs(title = "GDP oer capita of five countries", subtitle = "Mean of GDP per capita of their continent") 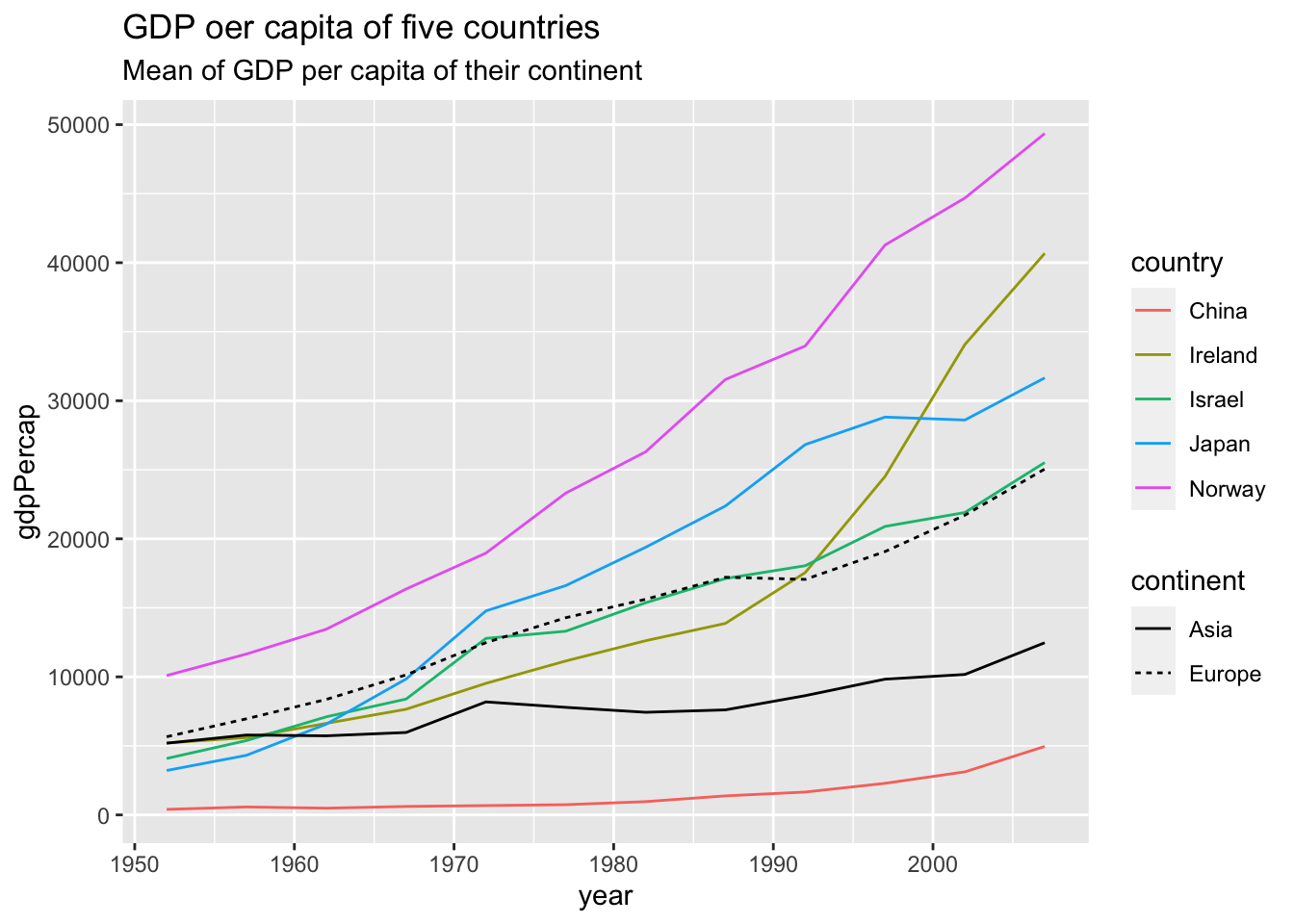
When you want to change the linetype manually, use scale_linetype_manual().
df %>%
group_by(continent, year) %>%
mutate(mean_by_continent = mean(gdpPercap)) %>%
ungroup() %>%
filter(country %in% c("Israel", "Japan", "Norway", "China", "Ireland")) %>%
ggplot(aes(x = year)) +
geom_line(aes(y = gdpPercap, color=country)) +
geom_line(aes(y = mean_by_continent, linetype=continent)) +
scale_linetype_manual(values = c("Asia" = "twodash", "Europe" = "longdash")) +
labs(title = "GDP oer capita of five countries", subtitle = "Mean of GDP per capita of their continent") 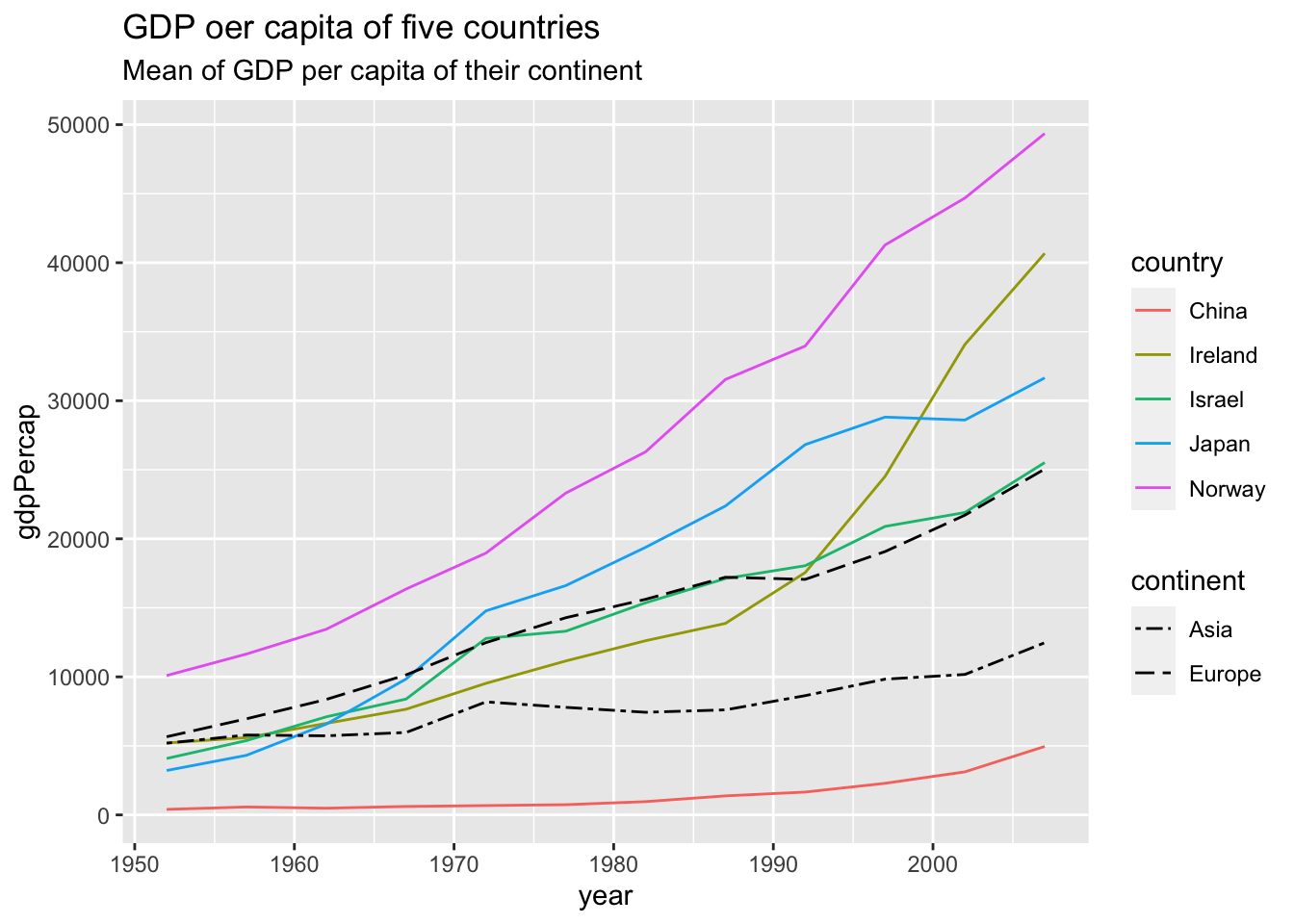
6.4 Appendix: Change colors, shapes, linetypes, etc. manually
Example: Default
df %>%
filter(country %in% c("Germany", "Japan", "United States")) %>%
ggplot() +
geom_line(aes(x = year, y = gdpPercap, color=country, linetype=continent))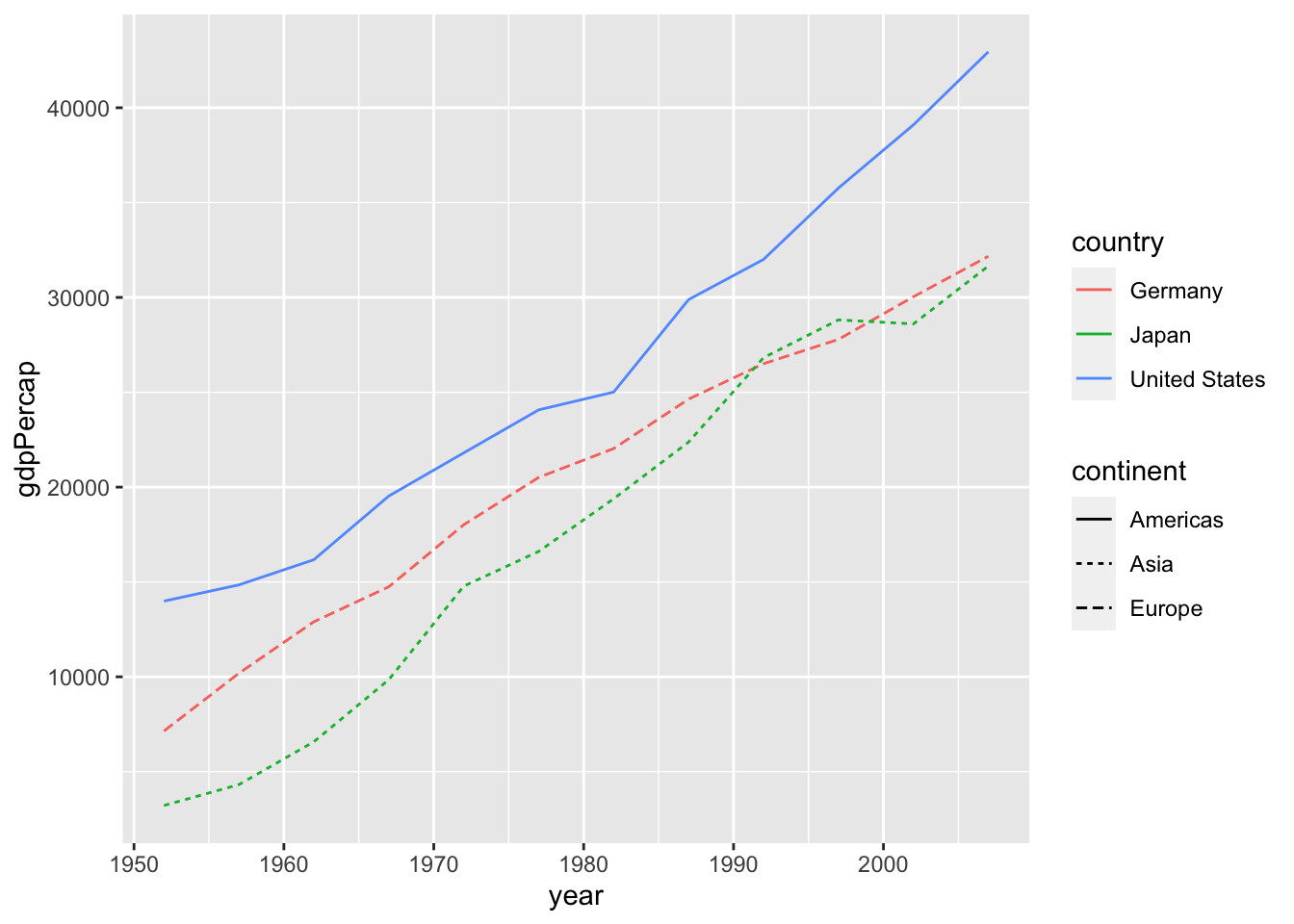
-
scale_color_manual: https://ggplot2-book.org/scale-colour.html- eg1: scale_colour_manual(values = c(“red”, “blue”, “green”))
- eg2: scale_colour_manual(values = c(“China” = “red”, “Japan” = “blue”, “Norway” = “green”))
- eg3: scale_colour_manual(values = scales::hue_pal()(3)) # default
- eg4: scale_colour_manual(values = scales::hue_pal(direction = -1)(3)) # reverse order
-
scale_fill_manual: similar toscale_color_manual -
scale_linetype_manual: https://ggplot2-book.org/scale-other.html?q=linetype#scale-linetype - `scale_shape_manual: https://ggplot2-book.org/scale-other.html?q=scale_shape_manual#scale-shape
- `scale_size: https://ggplot2-book.org/scale-other.html?q=size#scale-size
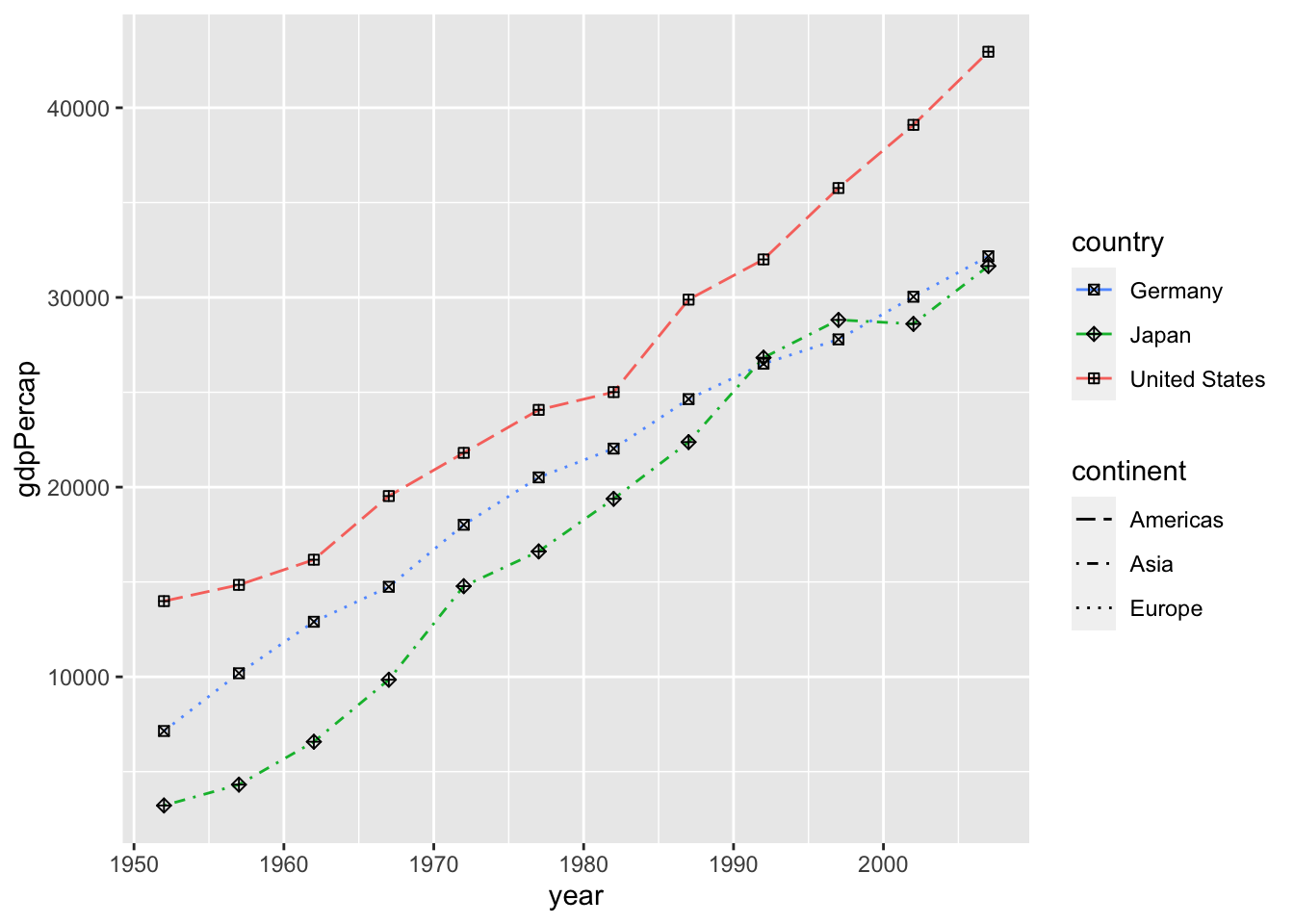
df %>%
filter(country %in% c("Germany", "Japan", "United States")) %>%
ggplot(aes(x = year, y = gdpPercap)) +
geom_line(aes(color=country, linetype=continent)) +
geom_point(aes(shape = country)) +
scale_colour_manual(values = scales::hue_pal(direction = -1)(3)) +
scale_linetype_manual(values = c("Europe" = "dotted", "Asia" = "dotdash", "Americas" = "longdash")) +
scale_shape_manual(values = c("Germany" = 7, "Japan" = 9, "United States" = 12))