2 R and R Studio
2.1 What is R?
2.1.1 R (programming language), Wikipedia
R is a programming language and free software environment for statistical computing and graphics supported by the R Foundation for Statistical Computing.
The R language is widely used among statisticians and data miners for developing statistical software and data analysis.
A GNU package, the official R software environment is written primarily in C, Fortran, and R itself (thus, it is partially self-hosting) and is freely available under the GNU General Public License.
2.2 Why R? – Responses by Hadley Wickham
2.2.1 r4ds: R is a great place to start your data science journey because
- R is an environment designed from the ground up to support data science.
- R is not just a programming language, but it is also an interactive environment for doing data science.
- To support interaction, R is a much more flexible language than many of its peers.
2.2.2 Why R today?
When you talk about choosing programming languages, I always say you shouldn’t pick them based on technical merits, but rather pick them based on the community. And I think the R community is like really, really strong, vibrant, free, welcoming, and embraces a wide range of domains. So, if there are like people like you using R, then your life is going to be much easier. That’s the first reason.
Interview: “Advice to Young (and Old) Programmers, H. Wickham”
2.3 What is RStudio? https://posit.com
RStudio is an integrated development environment, or IDE, for R programming.
2.3.1 R Studio (Wikipedia)
RStudio is an integrated development environment (IDE) for R, a programming language for statistical computing and graphics. It is available in two formats: RStudio Desktop is a regular desktop application while RStudio Server runs on a remote server and allows accessing RStudio using a web browser.
2.4 Installation of R and R Studio
2.4.1 R Installation
To download R, go to CRAN, the comprehensive R archive network. CRAN is composed of a set of mirror servers distributed around the world and is used to distribute R and R packages. Don’t try and pick a mirror that’s close to you: instead use the cloud mirror, https://cloud.r-project.org, which automatically figures it out for you.
A new major version of R comes out once a year, and there are 2-3 minor releases each year. It’s a good idea to update regularly.
2.4.2 R Studio Installation
Download and install it from http://www.rstudio.com/download.
RStudio is updated a couple of times a year. When a new version is available, RStudio will let you know.
2.5 R Studio
2.5.1 The First Step
- Start R Studio Application
- Top Menu: File > New Project > New Directory > New Project > Directory name or Browse the directory and choose the parent directory you want to create the directory
2.5.2 When You Start the Project
- Go to the directory you created
- Double click _‘Directory Name’.Rproj
Or,
- Start R Studio
- File > Open Project (or choose from Recent Project)
In this way the working directory of the session is set to the project directory and R can search releted files without difficulty (getwd(), setwd())
2.6 Posit Cloud
RStudio Cloud is a lightweight, cloud-based solution that allows anyone to do, share, teach and learn data science online.
2.6.1 Cloud Free
- Up to 15 projects total
- 1 shared space (5 members and 10 projects max)
- 15 project hours per month
- Up to 1 GB RAM per project
- Up to 1 CPU per project
- Up to 1 hour background execution time
2.6.2 How to Start Posit Cloud
- Go to https://posit.cloud/
- Sign Up: top right
- Email address or Google account
- New Project: Project Name
- R Console
2.7 Let’s Get Started
Start RStudio and create a project, or login to Posit Cloud and create a project.
2.7.1 The First Examples
Input the following codes into Console in the left bottom pane.
- The first two:
head(cars)
#> speed dist
#> 1 4 2
#> 2 4 10
#> 3 7 4
#> 4 7 22
#> 5 8 16
#> 6 9 10
str(cars)
#> 'data.frame': 50 obs. of 2 variables:
#> $ speed: num 4 4 7 7 8 9 10 10 10 11 ...
#> $ dist : num 2 10 4 22 16 10 18 26 34 17 ...- Two more:
summary(cars)
#> speed dist
#> Min. : 4.0 Min. : 2.00
#> 1st Qu.:12.0 1st Qu.: 26.00
#> Median :15.0 Median : 36.00
#> Mean :15.4 Mean : 42.98
#> 3rd Qu.:19.0 3rd Qu.: 56.00
#> Max. :25.0 Max. :120.00
plot(cars)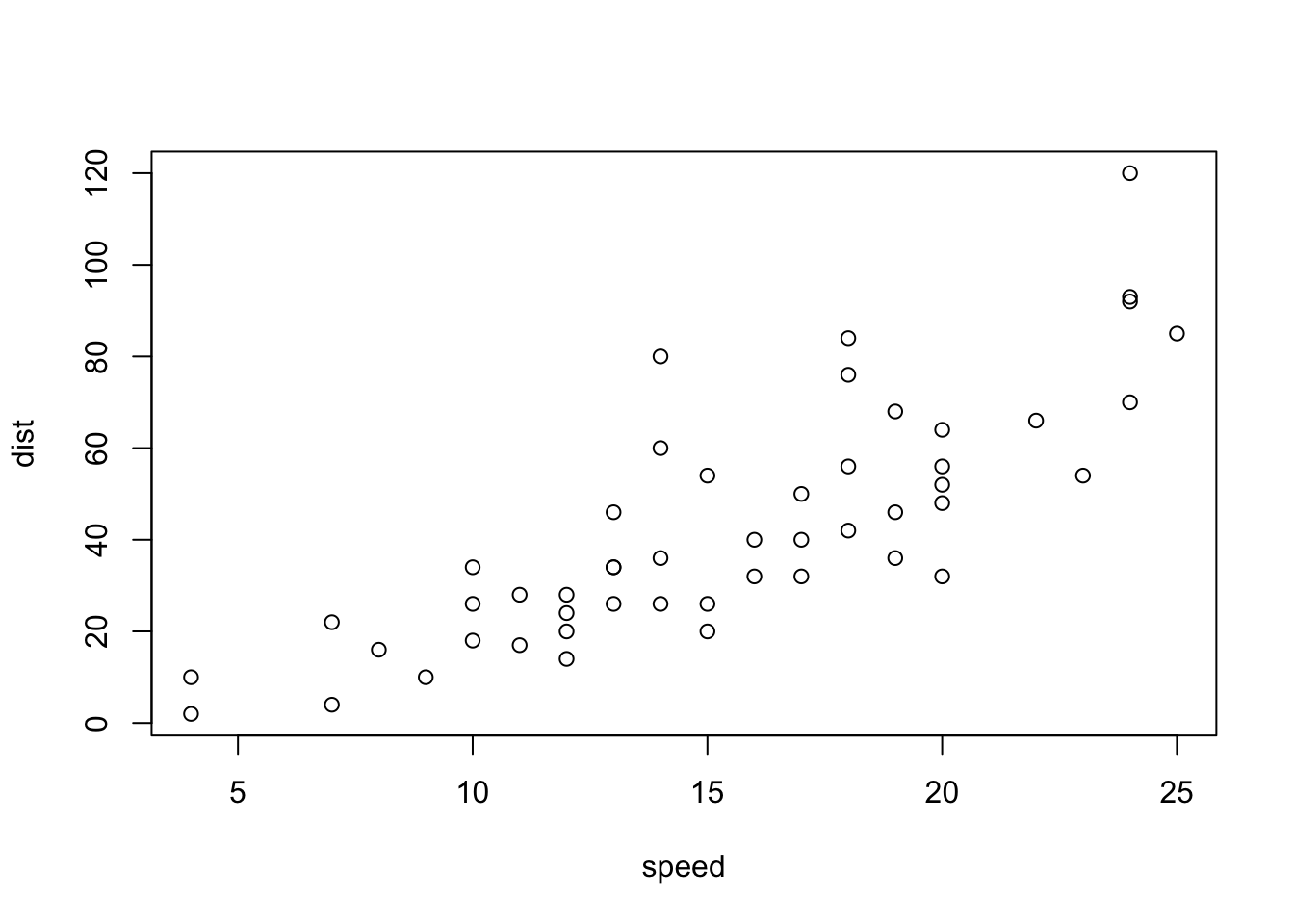
- And three more:
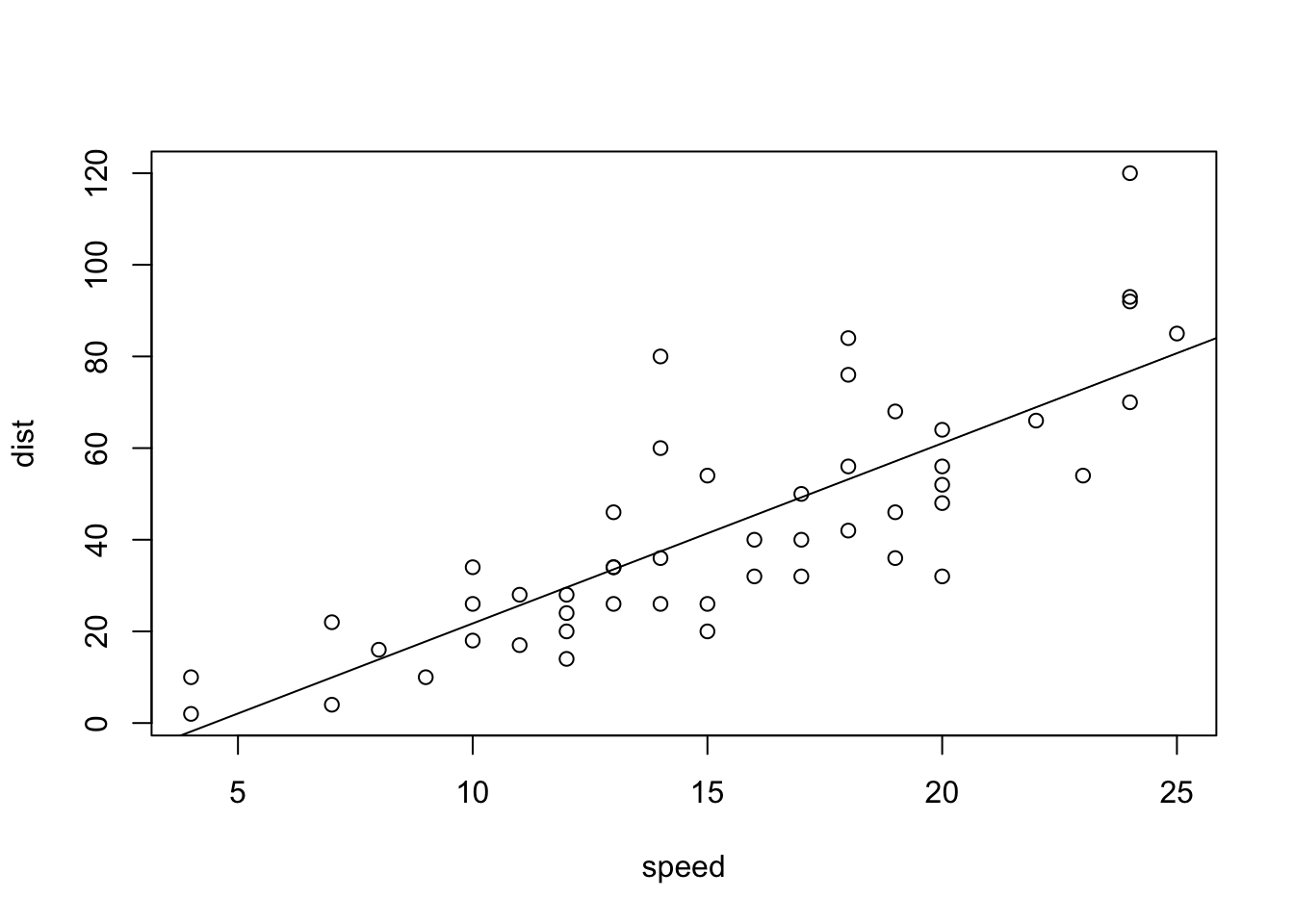
lm(cars$dist~cars$speed)
#>
#> Call:
#> lm(formula = cars$dist ~ cars$speed)
#>
#> Coefficients:
#> (Intercept) cars$speed
#> -17.579 3.932
summary(lm(cars$dist~cars$speed))
#>
#> Call:
#> lm(formula = cars$dist ~ cars$speed)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -29.069 -9.525 -2.272 9.215 43.201
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -17.5791 6.7584 -2.601 0.0123 *
#> cars$speed 3.9324 0.4155 9.464 1.49e-12 ***
#> ---
#> Signif. codes:
#> 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 15.38 on 48 degrees of freedom
#> Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
#> F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-122.7.1.1 Brief Explanation
cars: A pre-installed data which is a part of packagedatasets.head(cars): The first six rows of the pre-installed datacars.str(cars): The pre-installed datacarsdata structure.summary(cars): The summary of the pre-installed datacars.-
plot(cars): A scatter plot of the pre-installed datacars.plot(cars$dist~cars$speed)-
cars$dist,cars$[[2]],cars[,2]are same
abline(lm(cars$dist~cars$speed)): Add a regression line of a linear modellm(cars$dist~cars$speed): The equation of the regression linesummary(lm(cars$dist~cars$speed): The summary of the linear regression model
2.7.1.2 Histograms
cars is a data frame consisting of two columns, dist and speed.
hist(cars$dist)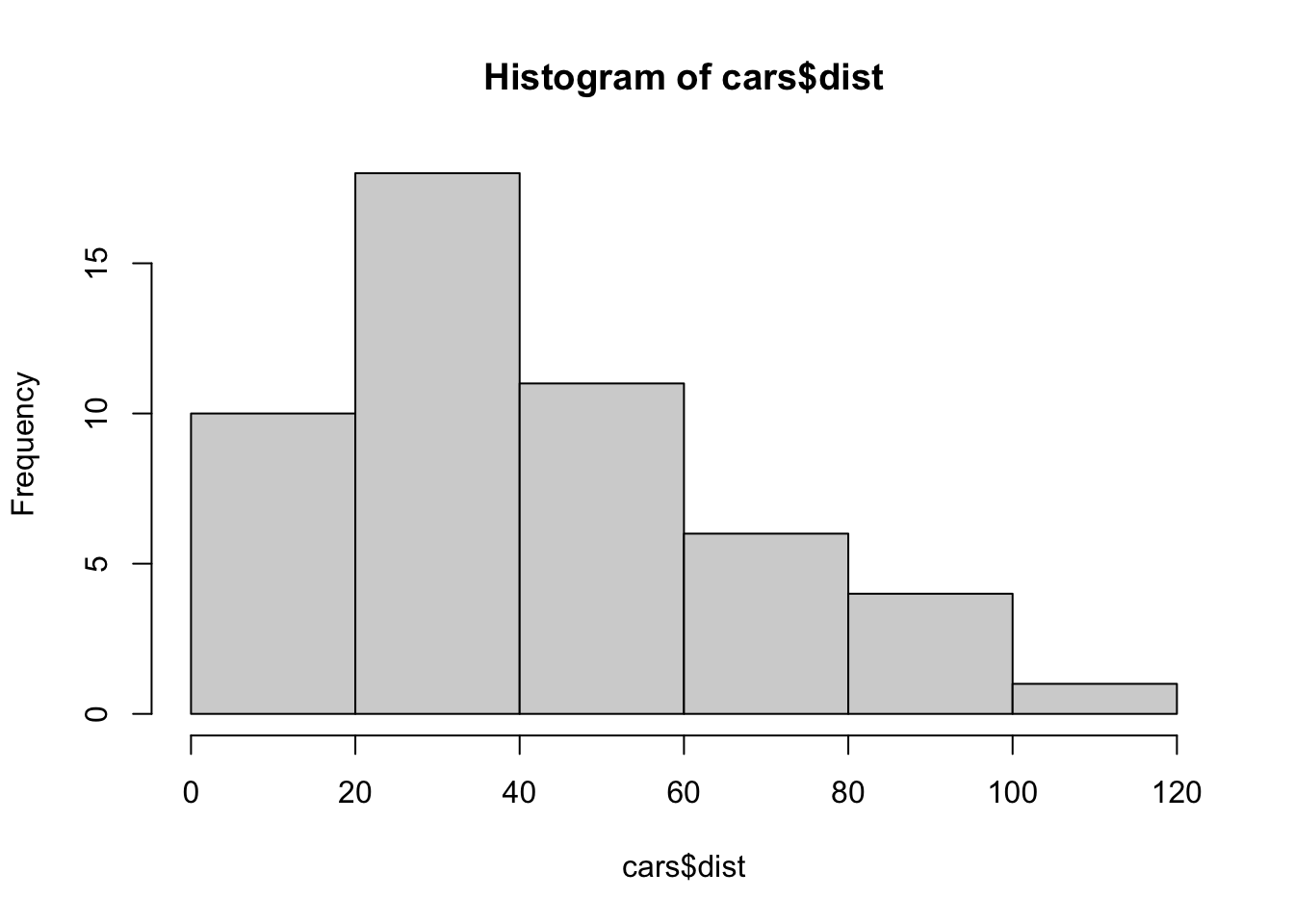
hist(cars$speed)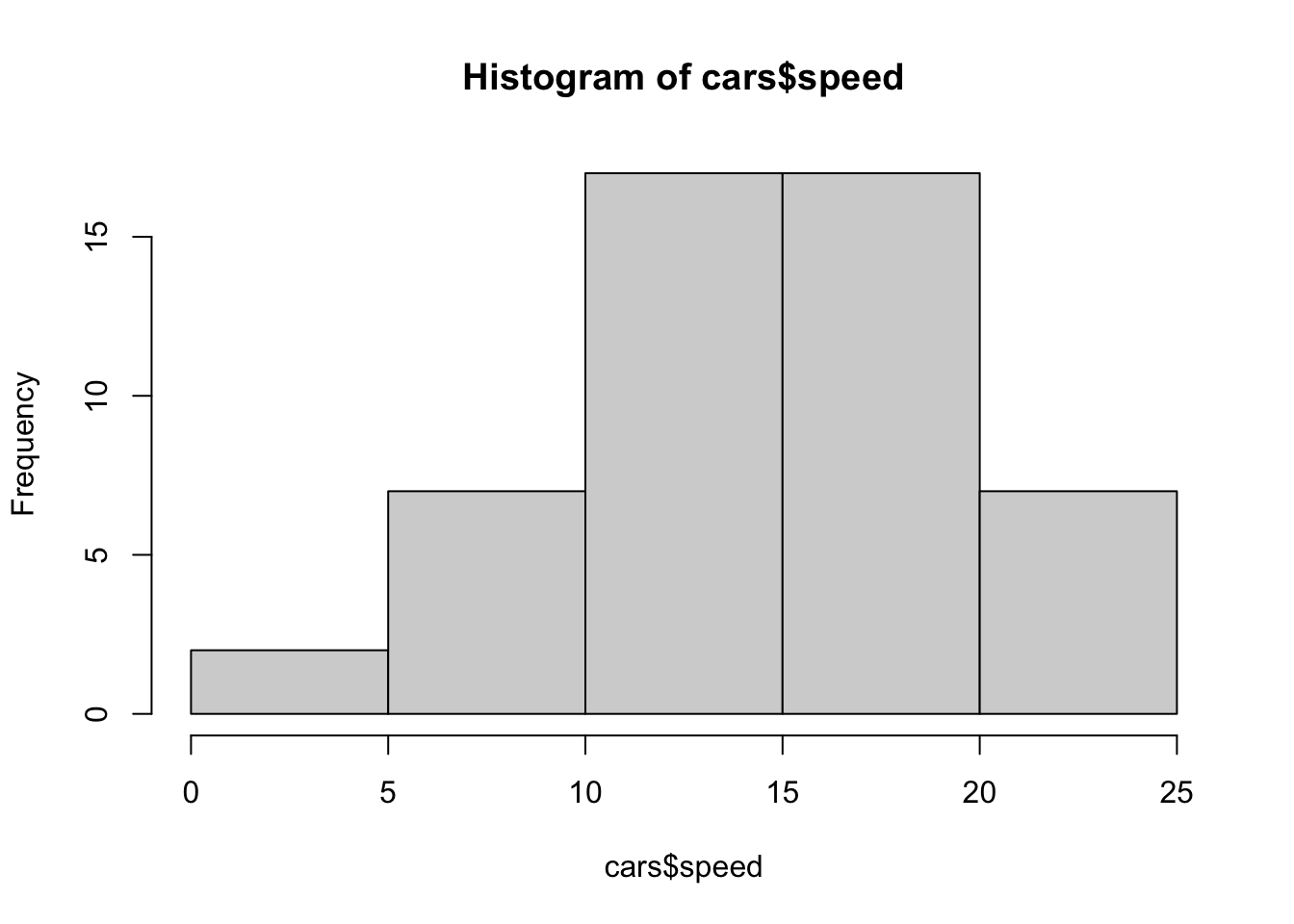
2.7.1.3 View and help
View(cars)-
?cars: same ashelp(cars) -
??cars: same as `help.search(“cars”)
2.7.1.4 datasets
datasets is a pre-installed datasets which contains a variety of datasets.
data()shows all data already attached and available.
By library(help = "datasets") and/or data(), you can see the complete list of data in datasets.
2.7.3 iris
head(iris)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
str(iris)
#> 'data.frame': 150 obs. of 5 variables:
#> $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
#> $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
#> $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
#> $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
#> $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
summary(iris)
#> Sepal.Length Sepal.Width Petal.Length
#> Min. :4.300 Min. :2.000 Min. :1.000
#> 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600
#> Median :5.800 Median :3.000 Median :4.350
#> Mean :5.843 Mean :3.057 Mean :3.758
#> 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100
#> Max. :7.900 Max. :4.400 Max. :6.900
#> Petal.Width Species
#> Min. :0.100 setosa :50
#> 1st Qu.:0.300 versicolor:50
#> Median :1.300 virginica :50
#> Mean :1.199
#> 3rd Qu.:1.800
#> Max. :2.500Can you plot?
plot(iris$Sepal.Length, iris$Sepal.Width)
2.8 Brief Introduction to R on RStudio
2.9 Set up
- Highly recommend to set the language to be “English”.
- Create “data” directory.
Sys.setenv(LANG = "en")
dir.create("./data")2.10 Three Ways to Run Codes
- Console - Bottom Left Pane
- We have run codes on the console already!
- R Script - pull-down menu under File
- R Notebook, R Markdown - pull down menu under File
2.11 R Script - Second Way to Run Codes
2.11.1 Examples: R Scripts in Moodle
basics.Rcoronavirus.R
- Copy a script in Moodle: {file name}.R
- In RStudio (create Project in RStudio) choose File > New File > R Script and paste it.
- Choose File > Save As, save with a name; e.g. {file names} (.R will be added automatically)
To run a code: at the cursor press Ctrl+Shift+Enter (Win) or Cmd+Shift+Enter (Mac).
- Top Manu: Help > Keyboard Short Cut Help contains many shortcuts.
- Bottom Right Pane: Check the files by selecting the
Filestab.
2.12 Practicum
Run the following and see what happens. You do not have to understand everything. Please guess what each code does.
2.12.1 R Scripts in Moodle
- basics.R
- coronavirus.R
- Copy a script in Moodle: {file name}.R
- In RStudio (Workspace in RStudio.cloud, Project in RStudio) choose File > New File > R Script and paste it.
- Choose File > Save with a name; e.g. {file names} (.R will be added automatically)
2.12.2 basics.R
The script with the outputs.
#################
#
# basics.R
#
################
# 'Quick R' by DataCamp may be a handy reference:
# https://www.statmethods.net/management/index.html
# Cheat Sheet at RStudio: https://www.rstudio.com/resources/cheatsheets/
# Base R Cheat Sheet: https://github.com/rstudio/cheatsheets/raw/main/base-r.pdf
# To execute the line: Control + Enter (Window and Linux), Command + Enter (Mac)
## try your experiments on the console
## calculator
3 + 7
### +, -, *, /, ^ (or **), %%, %/%
3 + 10 / 2
3^2
2^3
2*2*2
### assignment: <-, (=, ->, assign())
x <- 5
x
#### object_name <- value, '<-' shortcut: Alt (option) + '-' (hyphen or minus)
#### Object names must start with a letter and can only contain letter, numbers, _ and .
this_is_a_long_name <- 5^3
this_is_a_long_name
char_name <- "What is your name?"
char_name
#### Use 'tab completion' and 'up arrow'
### ls(): list of all assignments
ls()
ls.str()
#### check Environment in the upper right pane
### (atomic) vectors
5:10
a <- seq(5,10)
a
b <- 5:10
identical(a,b)
seq(5,10,2) # same as seq(from = 5, to = 10, by = 2)
c1 <- seq(0,100, by = 10)
c2 <- seq(0,100, length.out = 10)
c1
c2
length(c1)
#### ? seq ? length ? identical
(die <- 1:6)
zero_one <- c(0,1) # same as 0:1
die + zero_one # c(1,2,3,4,5,6) + c(0,1). re-use
d1 <- rep(1:3,2) # repeat
d1
die == d1
d2 <- as.character(die == d1)
d2
d3 <- as.numeric(die == d1)
d3
### class() for class and typeof() for mode
### class of vectors: numeric, charcters, logical
### types of vectors: doubles, integers, characters, logicals (complex and raw)
typeof(d1); class(d1)
typeof(d2); class(d2)
typeof(d3); class(d3)
sqrt(2)
sqrt(2)^2
sqrt(2)^2 - 2
typeof(sqrt(2))
typeof(2)
typeof(2L)
5 == c(5)
length(5)
### Subsetting
(A_Z <- LETTERS)
A_F <- A_Z[1:6]
A_F
A_F[3]
A_F[c(3,5)]
large <- die > 3
large
even <- die %in% c(2,4,6)
even
A_F[large]
A_F[even]
A_F[die < 4]
### Compare df with df1 <- data.frame(number = die, alphabet = A_F)
df <- data.frame(number = die, alphabet = A_F, stringsAsFactors = FALSE)
df
df$number
df$alphabet
df[3,2]
df[4,1]
df[1]
class(df[1])
class(df[[1]])
identical(df[[1]], die)
identical(df[1],die)
####################
# The First Example
####################
plot(cars)
# Help
? cars
# cars is in the 'datasets' package
data()
# help(cars) does the same as ? cars
# You can use Help tab in the right bottom pane
help(plot)
? par
head(cars)
str(cars)
summary(cars)
x <- cars$speed
y <- cars$dist
min(x)
mean(x)
quantile(x)
plot(cars)
abline(lm(cars$dist ~ cars$speed))
summary(lm(cars$dist ~ cars$speed))
boxplot(cars)
hist(cars$speed)
hist(cars$dist)
hist(cars$dist, breaks = seq(0,120, 10))2.13 Swirl: An interactive learning environment for R and statistics
- {
swirl} website: https://swirlstats.com - JHU Data Science in coursera uses
swirlfor exercises.
2.13.1 Swirl Courses
- R Programming: The basics of programming in R
- Regression Models: The basics of regression modeling in R
- Statistical Inference: The basics of statistical inference in R
- Exploratory Data Analysis: The basics of exploring data in R
You can install other swirl courses as well
- Swirl Courses Organized by Title
- Swirl Courses Organized by Author’s Name
-
Github: swirl courses
install_course("Course Name Here")
2.13.3 R Programming: The basics of programming in R
1: Basic Building Blocks 2: Workspace and Files 3: Sequences of Numbers
4: Vectors 5: Missing Values 6: Subsetting Vectors
7: Matrices and Data Frames 8: Logic 9: Functions
10: lapply and sapply 11: vapply and tapply 12: Looking at Data
13: Simulation 14: Dates and Times 15: Base Graphics 2.13.4 Recommended Sections in Order
1, 3, 4, 5, 6, 7, 12, 15, 14, 8, 9, 10, 11, 13, 2
- Section 2 discusses the directories and file systems of a computer
- Sections 9, 10, 11 are for programming
2.13.5 Controling a swirl Session
… <– That’s your cue to press Enter to continue
You can exit swirl and return to the R prompt (>) at any time by pressing the Esc key.
If you are already at the prompt, type bye() to exit and save your progress. When you exit properly, you’ll see a short message letting you know you’ve done so.
When you are at the R prompt (>):
- Typing skip() allows you to skip the current question.
- Typing play() lets you experiment with R on your own; swirl will ignore what you do…
- UNTIL you type nxt() which will regain swirl’s attention.
- Typing bye() causes swirl to exit. Your progress will be saved.
- Typing main() returns you to swirl’s main menu.
- Typing info() displays these options again.